We present the sTandalone Recorder of EXperiments T-REX, an open-source, OS-independent, user-friendly, scalable, and customizable web interface for recording (neuroscientific) experiments. Under the hood, T-REX handles data synchronization from multiple streams and ensures all data streams are recorded and saved to the user-specified location. By taking care of the overhead, the researcher can spend all their time recording experiments.
For a full description of the project, please check the preprint: T-Rex: sTandalone Recorder of EXperiments; An easy and versatile neural recording platform.
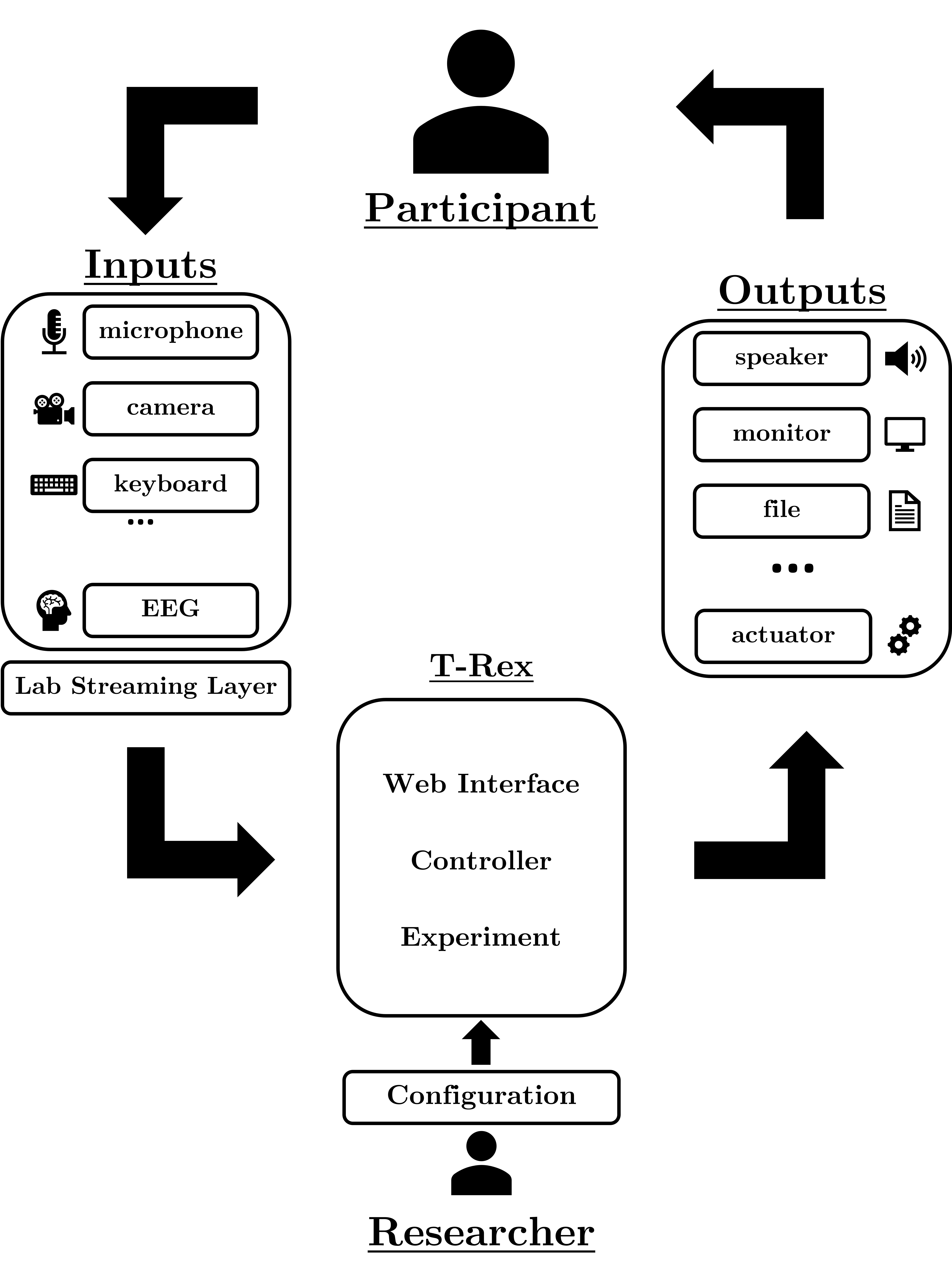
T-Rex acts as the middle man handling the experimental overhead for the researcher (Figure 1). When using T-Rex, the researcher can select an experiment by simply pressing a button on the main menu screen (Figure 2 A). When an experiment is selected, T-Rex checks the availability of all required input devices for the selected experiment and establishes a connection. For example, a movement-tracking experiment involves input from a hand-tracking device and the amplifier measuring the participants' neural activity. T-Rex will search for and connect to these devices. T-Rex will also start the experiment user interface (UI) that instructs the participant on what task to perform. It starts recording all data streams and saves them to a folder specified by the researcher. The synchronized data is saved inside this folder using the .xdf file format. Lastly, the UI prompts the participant on how the experiment went and returns to the Home screen, where the following experiment can be selected. The only action the researcher has to perform is to start the required device data stream(s) and choose the experiment from the Home screen.
The web interface is built using Bootstrap5 for the front end, and the Python Flask for the back-end.
The Controller, handling set up, start, and stop of the experiments, was built with Python 3.9 and requires a few dependencies found in the requirements.txt file located inside the folder resources of the repository. Data synchronization is implemented using Lab Streaming Layer.
T-Rex is compatible with Windows, macOS, and Linux. As T-Rex is lightweight, the hardware requirements for running it are defined mainly by the experiments it will execute.
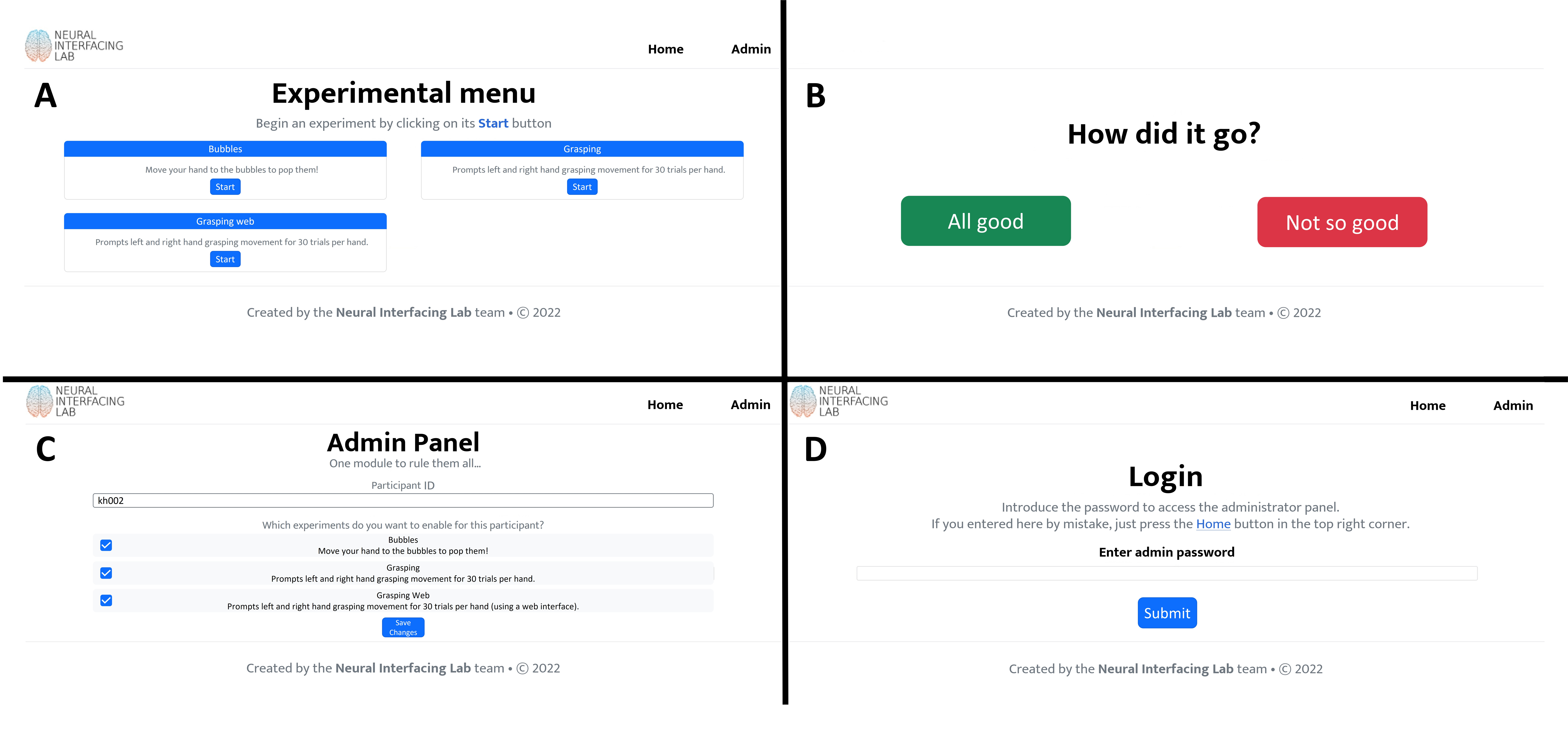
The Web interface includes four windows: Home, Experiment Feedback, Admin Login, and Admin Configuration (Figure 2).
The Home window (Figure 2 A) displays all the experiments in a grid. Experiments' cards' are shown on that grid with a title, description, and start button. When the button is pressed, the Controller executes a command that starts the selected experiment. The researcher defines and specifies the command in the experiment's configuration file. During the experiment, the web interface is on standby, awaiting the completion of the experiment.
Upon completion, the participant is redirected to the Experiment Feedback window where the question "How did the experiment go?" is prompted (Figure 2 B). The participants are required to select a feedback option to continue. This allows the researcher to save a brief experiment evaluation that quickly assesses the data quality in later analysis. In potential future applications, the participant might perform the experiments by themselves. Then this feedback is useful to flag the researcher to be aware of a potential loss in data quality during later analysis. The feedback is stored under the file name feedback.txt in the same folder as the most recent .xdf file (that contains the data recorded from the experiment).
The Admin Configuration provides the researcher with a closed environment where the participant identifier and a selection of all available experiments can be chosen. To access the Admin Configuration, the researcher must first log in using the password that is configured in the main configuration file (Figure 2 C). When logged in, the researcher can see the configuration of the active experimental session, composed of an alphanumeric participant identifier and their access to experiments. A list of all the experiments included in the platform is visible from this window, but only those with checked marks are visible to the participant. The changes on this window are only applied after pressing the "Save" button at the end of the page.
Figure 2: Screenshots of the main four windows in the Web interface. (A) The Home window contains all the experiments accessible to the user, shown in a grid configuration. (B) The Experiment Feedback window allows for obtaining feedback from the participants about their experience with the experiment. It is achieved through the green ("All good") and the red ("Not so good") buttons. The participants can only continue after pressing one of these buttons. (C) The Admin Login window allows access to the administration panel by entering the password. (D) The Admin Configuration window, where the administrator can create new participants and modify their access to experiments.To install T-Rex, you must have pre-installed the following:
Python 3.7 (or higher).Git.Check this for installation instructions.
We also advise using a virtual environment for running T-Rex. If this is the first time you hear about virtual environments, check this.
One of the many virtual environment tools is Conda. It comes in two flavors, Anaconda and Miniconda. The first is a comprehensive Python installation with plenty of tools to easily and efficiently manage integrated applications, packages, and environments without using the command line. On the other hand, Miniconda offers the conda command-line tool and only includes the basic dependencies required to get started. We prefer Miniconda, but if you're unsure which option to choose, check this comparison to assist you in making a decision.
This installation process is standard among different operating systems. There can always be slight differences, for example, in how paths are defined or, for different virtual environment managers, how to create and start the environment. In the following, we provide the basic ideas behind the installation of T-Rex and also give an example of how the installation process could look for different OS.
If you are not using any virtual environment management tool, first, you clone the repository and install the necessary packages to run the project. The requirements.txt file contains the python packages to install and is located in the resources folder inside the cloned t-rex repository you just cloned.
Windows:
git clone https://github.com/neuralinterfacinglab/t-rex.git
pip install -r t-rex\resources\requirements.txtLinux:
git clone https://github.com/neuralinterfacinglab/t-rex.git
pip install -r t-rex/resources/requirements.txtmacOS:
git clone https://github.com/neuralinterfacinglab/t-rex.git
pip install -r t-rex/resources/requirements.txtIf, on the other hand, you are using a virtual environment management tool, then you first need to create the virtual environment, clone the repository from GitHub, and install the necessary dependencies for the project. The requirements.txt file contains the python packages to install and is located in the resources folder inside the cloned t-rex repository you just cloned. If you decided to use Miniconda, then the following code can help you:
Windows:
git clone https://github.com/neuralinterfacinglab/t-rex.git
conda create --name trex python=3.7
conda activate trex
pip install -r t-rex\resources\requirements.txtLinux:
git clone https://github.com/neuralinterfacinglab/t-rex.git
conda create --name trex python=3.7
conda activate trex
pip install -r t-rex/resources/requirements.txtmacOS:
git clone https://github.com/neuralinterfacinglab/t-rex.git
conda create --name trex python=3.7
conda activate trex
pip install -r t-rex/resources/requirements.txtOne of the libraries pip installs is pylsl, which depends on liblsl and is typically installed automatically. However, for some Linux and macOS distributions, it is not included. There is no one-size-fits-all solution for the installation, so we provide instructions on detecting and fixing the problem. If while you execute T-Rex following the steps of the Execution guide, you encounter an error similar to this:
Error while loading shared libraries: liblsl64.so.X.XX.X: cannot open shared object file: No such file or directoryThen you probably need to install liblsl manually. If using conda this could be of help or using the releases.
After completing these steps, T-Rex is installed. To start the Web interface, check the Execution guide. If you want to try out the included example experiments you must install their requirements.
If you want to try out the included example experiments, their dependencies must first be installed:
Windows:
pip install -r t-rex\resources\requirements_for_example_exps.txtLinux:
pip install -r t-rex/resources/requirements_for_example_exps.txtmacOS:
pip install -r t-rex/resources/requirements_for_example_exps.txtTo test T-Rex you must:
-
open a console in the repositories' root directory
-
activate your python environment (if you are using one)
-
set a couple of environment variables required by Flask
FLASK_ENV=development FLASK_APP=flaskr
-
run flask
flask run
-
open your browser and check the address
http://127.0.0.1:5000
You can run these commands by hand or use the example scripts that we provide where we do all the above. However, you may need to modify these scripts because we make several assumptions. For example, we assume you use a virtual environment called trex and Conda to manage them. The good thing is that after modifying the scripts, they can quickly start T-Rex on your computer.
For some
call activate trex
set FLASK_ENV=development
set FLASK_APP=flaskr
start /max http://127.0.0.1:5000
flask runThis script can be found under the name run_windows.bat in the project's root folder. The first line activates the python environment, and you should check if this is the right way of starting the environment for your particular environment management tool. If you use conda, but it is not added to the PATH environment variable, then you should use the Anaconda Prompt to run the script.
Once modified, to start T-Rex, run the script in the console by typing:
run_windows.batsource activate trex
export FLASK_ENV=development
export FLASK_APP=flaskr
xdg-open http://127.0.0.1:5000
flask runThis script can be found under the name run_linux.sh in the project's root folder. The first line activates the python environment; you should check if this is the right way of starting the environment for your particular environment management tool. The xdg-open command opens the URL http://127.0.0.1:5000 in the default browser. However, it is not present in all Linux distributions by default and may not be available for distributions such as Arch Linux, Gentoo, and Slackware. In that case, you can try alternatives like gnome-open or kde-open, which are specific to Gnome and KDE desktop environments.
Execution permissions must be provided for this script. Open a console in the root project directory and type:
chmod +x run_linux.shOnce modified, to start T-Rex, run the script in the console by typing:
./run_linux.shconda activate trex
export FLASK_ENV=development
export FLASK_APP=flaskr
open http://127.0.0.1:5000
flask runThis script can be found under the name run_macos.sh in the project's root folder. The first line activates the python environment; you should check if this is the right way of starting the environment for your particular environment management tool. Execution permissions must be provided for this script. Open a console in the root project directory and type:
chmod +x run_macos.shOnce modified, to start T-Rex, run the script in the console by typing:
source run_macos.shAfter manually running the commands or modifying and executing the start scripts for your particular OS, a new tab should appear in your default browser. If a new tab is not there, type the address http://127.0.0.1:5000, which is the default URL the Flask development server runs on. The Home page (Figure 2 A) should be visible now.
You can start one of the experiments from the web interface to test that everything is working correctly. The current environment is configured as development, so a testing stream named DBG01 will start automatically, allowing you to try the system. Important: Before recording actual experiments, turn the debug mode off by setting the debug flag in the file resources/config.yaml to False.
The following steps describe how to add a new experiment from scratch to T-Rex:
- Create the experiment folder inside the directory
./exp_module/experiments/. - Create the experiment configuration file (
config.yaml) inside the new folder. - Adjust the fields to the specific experiment.
After completing these initial steps, the experiment is not visible as a security measure from the Home window. To make it visible, the researcher must access the Admin Configuration window and select the corresponding check mark. After pressing the Save button, it should appear on the Home window and be executed by clicking on its respective button.
Some parameters need to be checked when porting an already-configured version of T-Rex to a different OS. For example, the command used on Windows to start a python experiment is defined as follows:
command: python .\exp_module\experiments\example\example_experiment.pyHowever, when used on Unix or Unix-like systems, the definition changes to the following:
command: python ./exp_module/experiments/example/example_experiment.pyThe difference comes because / is the path separator on Unix and Unix-like systems, and Microsoft uses \.
There might be other scenarios where the parameter command might differ between OS, so we recommend revising each experiment configuration file when porting the platform to a different OS.
Even so, incorporating new experiments into T-Rex is seamless. Several experiments can be quickly integrated into the platform with just a few easy steps.
We have included three different example experiments to provide a more realistic view of how to use T-Rex. The examples can also serve as a quick start for researchers to create new experiments or adapt the ones included.
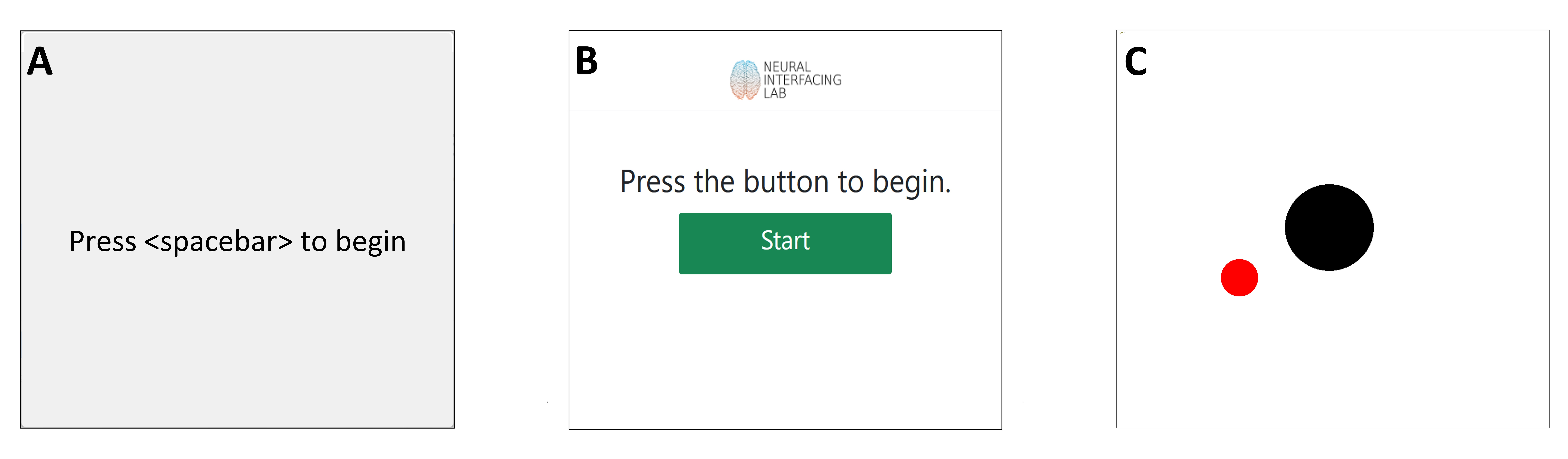
Figure 3: User interfaces for the three Use cases experiments included. (A) Grasping, simple text-based experiment built using the Python package Tkinter. (B) Grasping Web experiment, re-implementation of the Grasping experiment as a Single Page Application (SPA) to allow its execution on any device with access to a web browser. (C) 3D hand tracking experiment, the hand tracking is performed using the LeapMotion controller, and the experiment is implemented in Python using the package Tkinter.This experiment is a simple text-based instruction for a grasping task (Figure 3 A). The participant is prompted by text in a Python tkinter window to open and close either their left or right hand continuously. The experiment requires neural data as the input device and generates a StreamOutlet to send markers that inform about the start and end of the experiment and of the trials. The neural data is acquired from a stream with name=Micromed, type=EEG, source\_id=micm01. These values are all set by the user. As T-Rex will search for all three options (name, type, or source_id), only one needs to be provided. Therefore, the option under device_inputs in grasping\config.yaml is set to eeg (case-insensitive). Next, the Marker StreamOutlet that the experiment itself will generate has source\_id=emuidw22. When the Experiment class runs the experiment command (command field in grasping\config.yaml) it will search for these streams. Therefore, the exp_outlet field is set to emuidw22. Finally, since the grasping experiment is Python-based, the command should use Python to call the script with the command: python .\exp_module\experiments\grasping\grasping.py.
When these options are set, the experiment is ready to go and can be started by pressing the start button on the Home window. The Tk window opens and waits for the spacebar to be pressed. Once pressed, the experiment starts and is locked on the topmost position upon completion. When the experiment is finished and closed (i.e., the command call ends and returns to the Experiment class), the Experiment instance stops the recording and saves the data.
From the Web Interface, participants and researchers can access the main functionalities of T-Rex, allowing the potential execution of the system on a headless server, different from the interface that interacts with the participants. The system that runs T-Rex could create a local network that serves the experiments to other devices (tablets, laptops, smartphones), removing the need for an active internet connection which could render the execution unsafe.
To illustrate this paradigm, we created the grasping web experiment, which mimics the behavior of the "grasping" experiment but in a web format (Figure 3 B). Given that this experiment consists of a Single Page Application (SPA), it can be wholly executed on any device with access to a web browser, like laptops, tablets, and smartphones. The grasping web experiment also illustrates options other than a Tk window for experimenting.
For constructing the experiment, we used HTML, CSS (using Bootstrap5 for the responsiveness and other visual aspects), and JavaScript for the behavior. The device input is the same as in the Tk implementation and the StreamOutlet containing the markers. Thus, the device_inputs and exp_outlet are the same. The difference is in the command executed to start the experiment. In this case, start .\exp_module\experiments\graspingWeb\index.html| is used.
Once the experiment is started on the Home window, the Experiment instance opens another tab on the browser displaying the "grasping_web" experiment. The experiment starts when the participant presses the green "Start" button. When it is finished, the participant or researcher is prompted to press a red button to close the experiment. The GraspingWeb command call is finished at button-press and returns to the Experiment instance, stopping the recording and saving the data.
Lastly, we included a 3D hand-tracking experiment, where the goal is to hold the cursor (a black circle) on the target (a red circle). The cursor can be moved in 3d, where the third dimension controls the size of the circle (Figure 3 C). In this case, the hand tracking is done by the LeapMotion controller. We provided a .exe that reads the data from the tracker and sends it to an LSL StreamOutlet with name=LeapLSL, type=Coordinates, and source\_id=LEAPLSL01. In addition to the hand-tracking information, we also need neural activity, for which we use the same StreamOutlet described in case 2. Lastly, the experiment is implemented in a Python Tk window and generates a marker Stream similar to the streams defined in the previous use case with Source\_id=BUBBLE01. Thus, to set up the configuration for this experiment, we set command to python .\exp_module\experiments\Bubbles\bubbles.py, exp_outlet to BUBBLE01 and device_inputs to LEAPLSL01 (the tracking information stream) and eeg (the neural data stream). To run the experiment, the researcher should start the device stream before the experiment is started in the Home screen. (i.e., run the .exe first).
The collaborators of this project are: Joaquín Amigó-Vega, Maarten C. Ottenhoff, Maxime Verwoert, Pieter Kubben, and Christian Herff.
We want to thank also the members of the Neural Interfacing Lab.