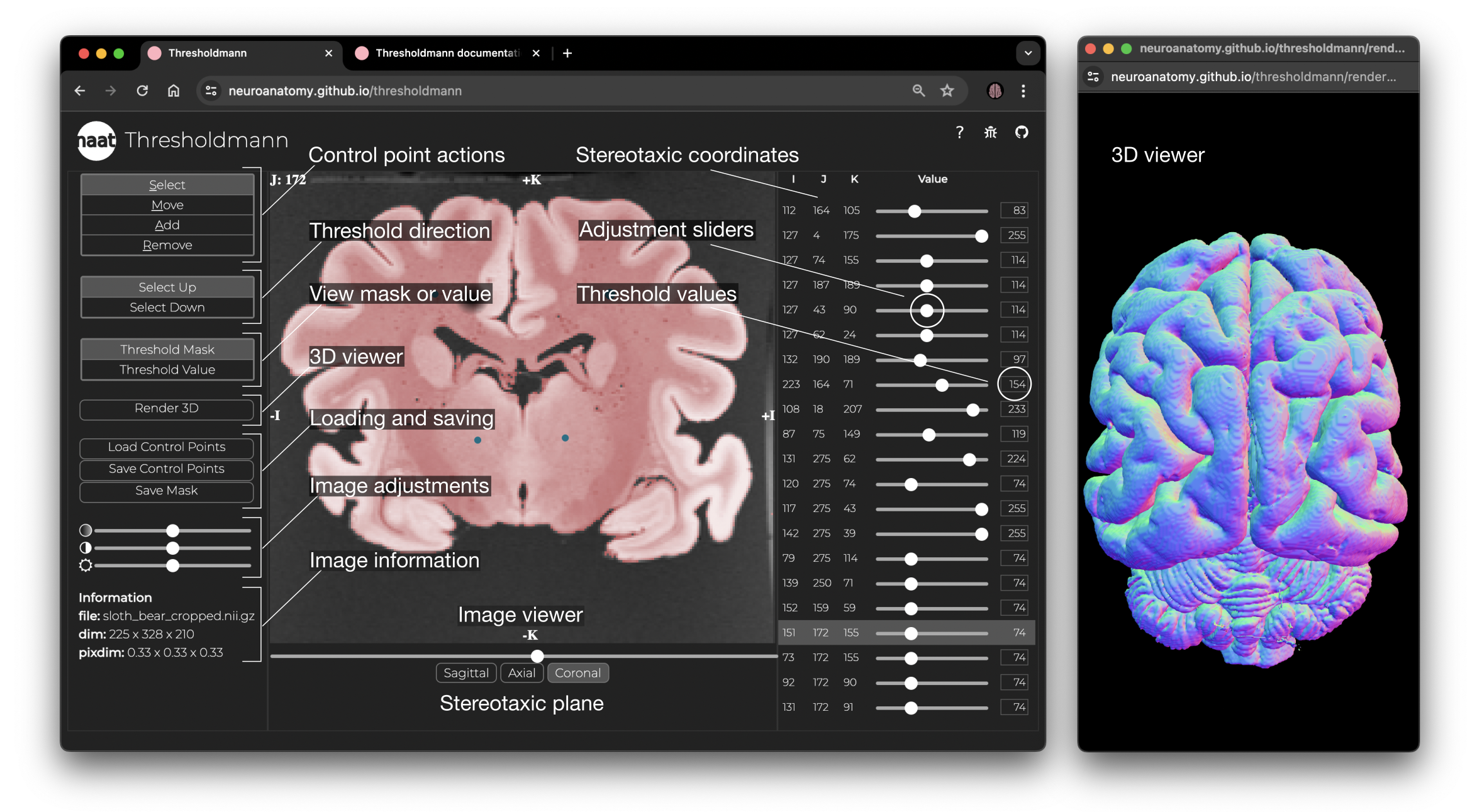
A Web tool for interactively creating adaptive thresholds to segment MRI data.
Katja Heuer, Nicolas Traut & Roberto Toro, November 2023
Simply drag & drop your MRI file for display in an interactive stereotaxic viewer. Move and add a control point and grow or shrink your selection from there.
You can later re-select and adjust control points or delete them. Once you are happy with your selection, you can download the control points (json) and you can download the mask (.nii.gz).
Brain extraction and segmentation are required for most analyses of neuroimaging data. Obtaining appropriate masks can be particularly difficult in non-human brain imaging, as standard automatic tools struggle with the surrounding muscle tissue, skull, and strong luminosity gradients. A simple interactive threshold is intuitive and fast to apply, and can often provide a rather good initial guess. However, because of luminosity gradients, the threshold that works for one brain region is likely to fail in another.
Thresholdmann is an open source Web tool for the interactive application of space-varying thresholds to Nifti volumes. No download or installation are required and all processing is done on the user’s computer. Nifti volumes are dragged and dropped onto the Web app and become available for visual exploration in a stereotaxic viewer. A space-varying threshold is then created by setting control points, each with their own local threshold. Each point can be repositioned or removed, and each local threshold can be adjusted in real time using sliders or entering their values numerically. The threshold direction can be switched to allow segmentation of the structure of interest in different imaging modalities, such as T1 and T2 weighted contrasts. The opacity of the mask and the brightness and contrast of the MRI image can be adjusted via sliders. A 3D model of the thresholded mask can be computed to inspect the result in an interactive 3D render. Finally, the thresholded mask, the space varying threshold and the list of control points can be saved for later use in scripted workflows, able to reproduce the thresholded volume from the original data.
Thresholdmann complements the variety of existing brain segmentation tools, providing an easy interface to manually control the segmentation on a local scale across different brain imaging modalities and image contrast gradients. The masks produced by Thresholdmann can serve as a starting point for more detailed manual editing using tools such as BrainBox or ITK Snap. This interactive approach is especially valuable for non-human brain imaging data, where automatic approaches often require extensive manual adjustment anyway. We have used Thresholdmann successfully to create initial brain masks for a variety of vertebrate brains – including many non-human primate datasets (Heuer et al. 2019, Magielse et al. 2023) – as well as developmental data. Small Web tools, such as Thresholdmann or Reorient (https://neuroanatomy.github.io/reorient), focused on solving a single problem, can become helpful additions to the methodological toolbox of neuroimagers.
A description of a typical workflow can be found in the doc.