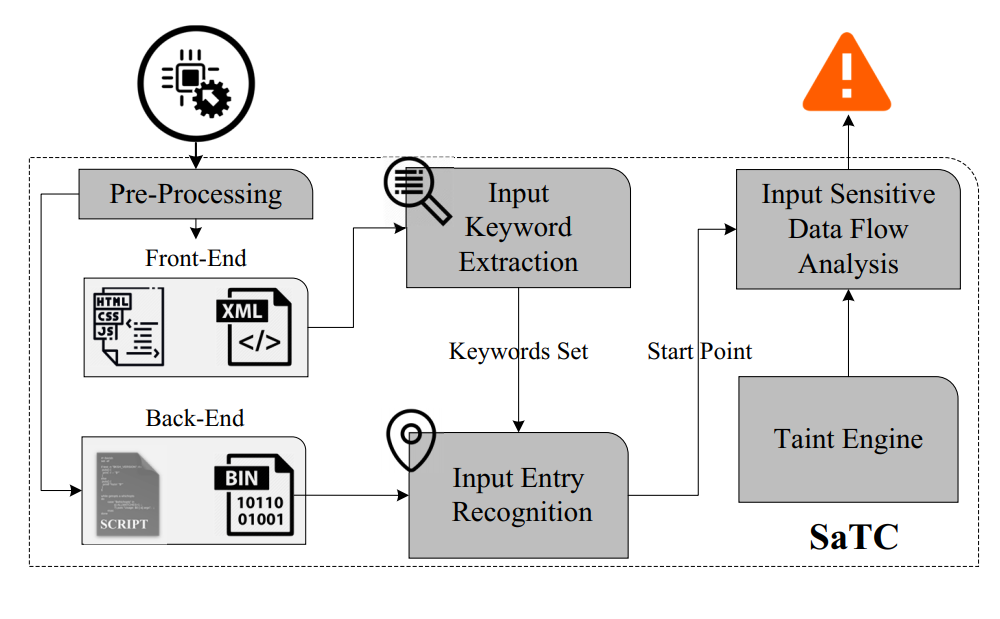
A prototype of Shared-keywords aware Taint Checking(SaTC), a static analysis method that tracks user input between front-end and back-end for vulnerability discovery effectively and efficiently.
We present our approach in the following research paper accepted at the 30th USENIX Security Symposium:
Sharing More and Checking Less: Leveraging Common Input Keywords to Detect Bugs in Embedded Systems [PDF]
We provide a usable Docker environment and a Dockerfile that can be used to build Docker images.
# Get image from Docker hub
docker pull smile0304/satc
# Run SaTC (Need to add mapping directory by yourself)
docker run -v <mapping>:<mapping> -it smile0304/satc# Cd SaTC code directory
cd SaTC
# Use Dockerfile to build docker image
docker build . -t satc
# Run SaTC (Need to add mapping directory by yourself)
docker run -v <mapping>:<mapping> -it satcUsage: satc.py [-h] -d /root/path/_ac18.extracted -o /root/output
[--ghidra_script {ref2sink_cmdi,ref2sink_bof,share2sink,ref2share,all}]
[--save_ghidra_project] --taint_check
[-b /var/ac18/bin/httpd | -l 3]
Arguments:
-h, --help Show help in details
-d /root/path/_ac18.extracted, --directory /root/path/_ac18.extracted
File system uncompressed from firmware
-o /root/output, --output /root/output
Directory result saved
--ghidra_script {ref2sink_cmdi,ref2sink_bof,share2sink,ref2share,all}
(Option) Specify the Ghidra script to be used. If you use the `all` command, the three scripts `ref2sink_cmdi`,`ref2sink_bof` and `ref2share` will be run at the same time
--ref2share_result /root/path/ref2share_result
(Option) When running the `share2sink` Ghidra script, you need to use this parameter to specify the output result of the `ref2share` script
--save_ghidra_project (Option) Save the ghidra project generated during analysis
--taint_check (Option) Use taint analysis engine for analysis
-b /var/ac18/bin/httpd, --bin /var/ac18/bin/httpd OR `-b httpd` , `--bin httpd`
(Option) Used to specify the program to be analyzed, if not specified, SaTC will leverage the built-in algorithm to match targeted bin
-l num, --len num (Option) To set the top N programs to be defined as the border bins in our matching results[Default value is 3]
ref2sink_cmdi: The script to discover the paths of the command injection type sink function from the reference of the given shared-keywords.ref2sink_bof: The script to discover the paths of the buffer overflow type sink function from the reference of the given shared-keywords.ref2share: This script to find parameters in shared data handling functions, such asnvram_set,setenvor other similar functions. Need to be used in conjunction with share2sink.share2sink: This script is corresponding toref2share, such asnvram_get,getenvor other functions. Need to be used in conjunction withref2share, and the input of this script is the output from theref2sharescript.
Directory structure:
|-- ghidra_extract_result
| |-- httpd
| |-- httpd
| |-- httpd_ref2sink_bof.result
| |-- httpd_ref2sink_cmdi.result
| |-- httpd_ref2sink_cmdi.result-alter2
|-- keyword_extract_result
| |-- detail
| | |-- API_detail.result
| | |-- API_remove_detail.result
| | |-- api_split.result
| | |-- Clustering_result_v2.result
| | |-- File_detail.result
| | |-- from_bin_add_para.result
| | |-- Not_Analysise_JS_File.result
| | |-- Prar_detail.result
| | |-- Prar_remove_detail.result
| |-- info.txt
| |-- simple
| |-- API_simple.result
| |-- Prar_simple.result
|-- result-httpd-ref2sink_cmdi-ctW8.txtNeed to follow such important directories:
- keyword_extract_result/detail/Clustering_result_v2.result : The match of front-end keywords in bin. Input for the
Input Entry Recognitionmodule - ghidra_extract_result/{bin}/* : Analysis result of ghidra script. Input for
Input Sensitive Taint Analysisemodule - result-{bin}-{ghidra_script}-{random}.txt: taint analysis result
Other directories:
|-- ghidra_extract_result # ghidra looks for the analysis results of the function call path, enabling the `--ghidra_script` option will output the directory
| |-- httpd # Each bin analyzed will generate a folder with the same name
| |-- httpd # Bin being analyzed
| |-- httpd_ref2sink_bof.result # Locate BoF sink function path
| |-- httpd_ref2sink_cmdi.result # Locate CmdI sink function path
|-- keyword_extract_result # Keyword extraction results
| |-- detail # Front-end keyword extraction results (detailed analysis results)
| | |-- API_detail.result # Detailed results of the extracted API
| | |-- API_remove_detail.result # API information filtered out
| | |-- api_split.result # Matching API results
| | |-- Clustering_result_v2.result # Detailed matching results
| | |-- File_detail.result # Keywords extracted from each file
| | |-- from_bin_add_para.result # Share-keywords generated during binary matching
| | |-- Not_Analysise_JS_File.result #Igored JS files by common lib matching
| | |-- Prar_detail.result # Detailed results of extracted Prarmeters
| | |-- Prar_remove_detail.result # Detailed results of filtered Prarmeters
| |-- info.txt # Record processing time and other information
|-- result-httpd-ref2sink_cmdi-ctW8.txt # a typical result file that enable `--taint-check` and `--ghidra_script` optionsYou should download dataset from SaTC_dateset.zip.
1.To discover command injection and buffer overflow bugs in D-Link 878
python satc.py -d /home/satc/dlink_878 -o /home/satc/res --ghidra_script=ref2sink_cmdi --ghidra_script=ref2sink_bof --taint_check2.To discover command injection bugs in specific target prog.cgi of D-Link 878
python satc.py -d /home/satc/dlink_878 -o /home/satc/res --ghidra_script=ref2sink_cmdi -b prog.cgi --taint_check3.To discover command injection bugs in multi-bin of D-Link 878, setting input data in prog.cgi and sink functions in rc
python satc.py -d /home/satc/dlink_878 -o /home/satc/res --ghidra_script=ref2share -b prog.cgi
python satc.py -d /home/satc/dlink_878 -o /home/satc/res --ghidra_script=share2sink --ref2share_result=/home/satc/res/ghidra_extract_result/prog.cgi/prog.cgi_ref2share.result -b rc --taint_check