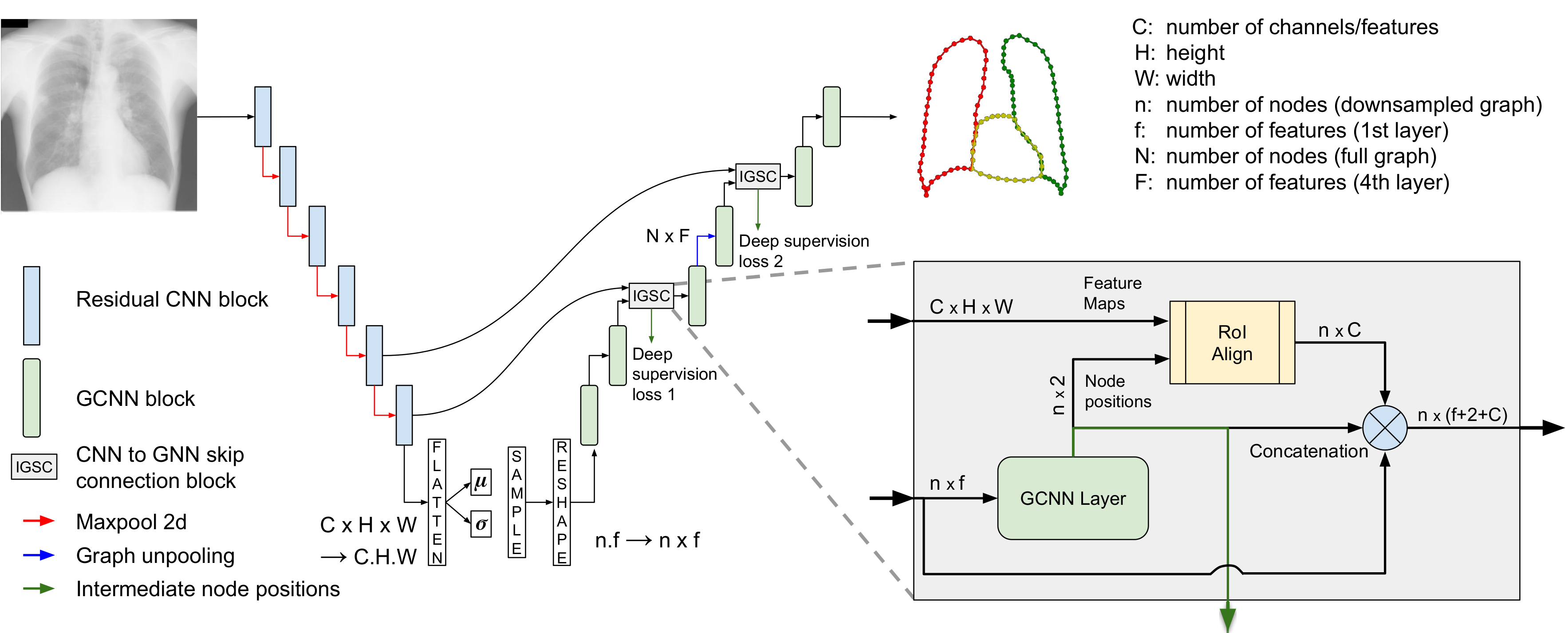
HybridGNet - Improving anatomical plausibility in image segmentation via hybrid graph neural networks: applications to chest x-ray image analysis
Nicolás Gaggion¹, Lucas Mansilla¹, Candelaria Mosquera²³, Diego Milone¹, Enzo Ferrante¹
¹ Research Institute for Signals, Systems and Computational Intelligence (sinc(i)), FICH-UNL, CONICET, Ciudad Universitaria UNL, Santa Fe, Argentina. ² Hospital Italiano de Buenos Aires, Buenos Aires, Argentina ³ Universidad Tecnológica Nacional, Buenos Aires, Argentina
Find it here! https://huggingface.co/spaces/ngaggion/Chest-x-ray-HybridGNet-Segmentation
IEEE TMI: https://doi.org/10.1109%2Ftmi.2022.3224660
Arxiv: https://arxiv.org/abs/2203.10977
Citation:
@article{Gaggion_2022,
doi = {10.1109/tmi.2022.3224660},
url = {https://doi.org/10.1109%2Ftmi.2022.3224660},
year = 2022,
publisher = {Institute of Electrical and Electronics Engineers ({IEEE})},
author = {Nicolas Gaggion and Lucas Mansilla and Candelaria Mosquera and Diego H. Milone and Enzo Ferrante},
title = {Improving anatomical plausibility in medical image segmentation via hybrid graph neural networks: applications to chest x-ray analysis},
journal = {{IEEE} Transactions on Medical Imaging}
}
For the old version of the code, check on Tags
Paper: https://link.springer.com/chapter/10.1007%2F978-3-030-87193-2_57
First create the anaconda environment:
conda env create -f environment.yml
Activate it with:
conda activate torch
In case the installation fails, you can build your own enviroment.
Conda dependencies:
-PyTorch 1.10.0
-Torchvision
-PyTorch Geometric
-Scipy
-Numpy
-Pandas
-Scikit-learn
-Scikit-image
Pip dependencies:
-medpy==0.4.0
-opencv-python==4.5.4.60
Download the datasets from the official sources (check Datasets/readme.txt) and run the corresponding preprocessing scripts.
A new dataset of landmark annotations was released jointly with this work. Available at https://github.com/ngaggion/Chest-xray-landmark-dataset
Download the weights from here: https://drive.google.com/drive/folders/1YcmT8JzdtNuaWVqhv8Zfm00lF47w0eU5
For more information about the MultiAtlas baseline, check Lucas Mansilla's repository: https://github.com/lucasmansilla/multiatlas-landmark