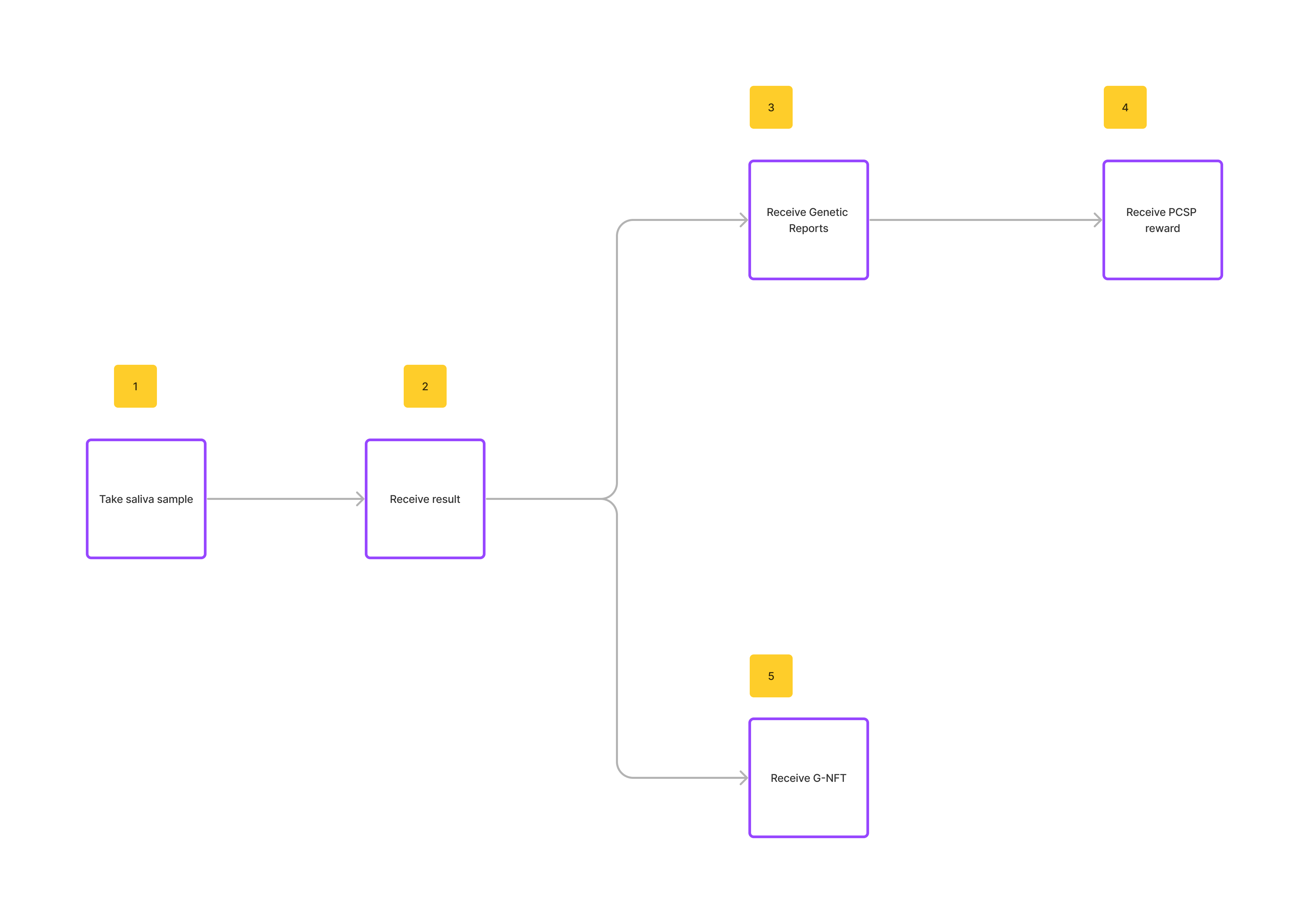
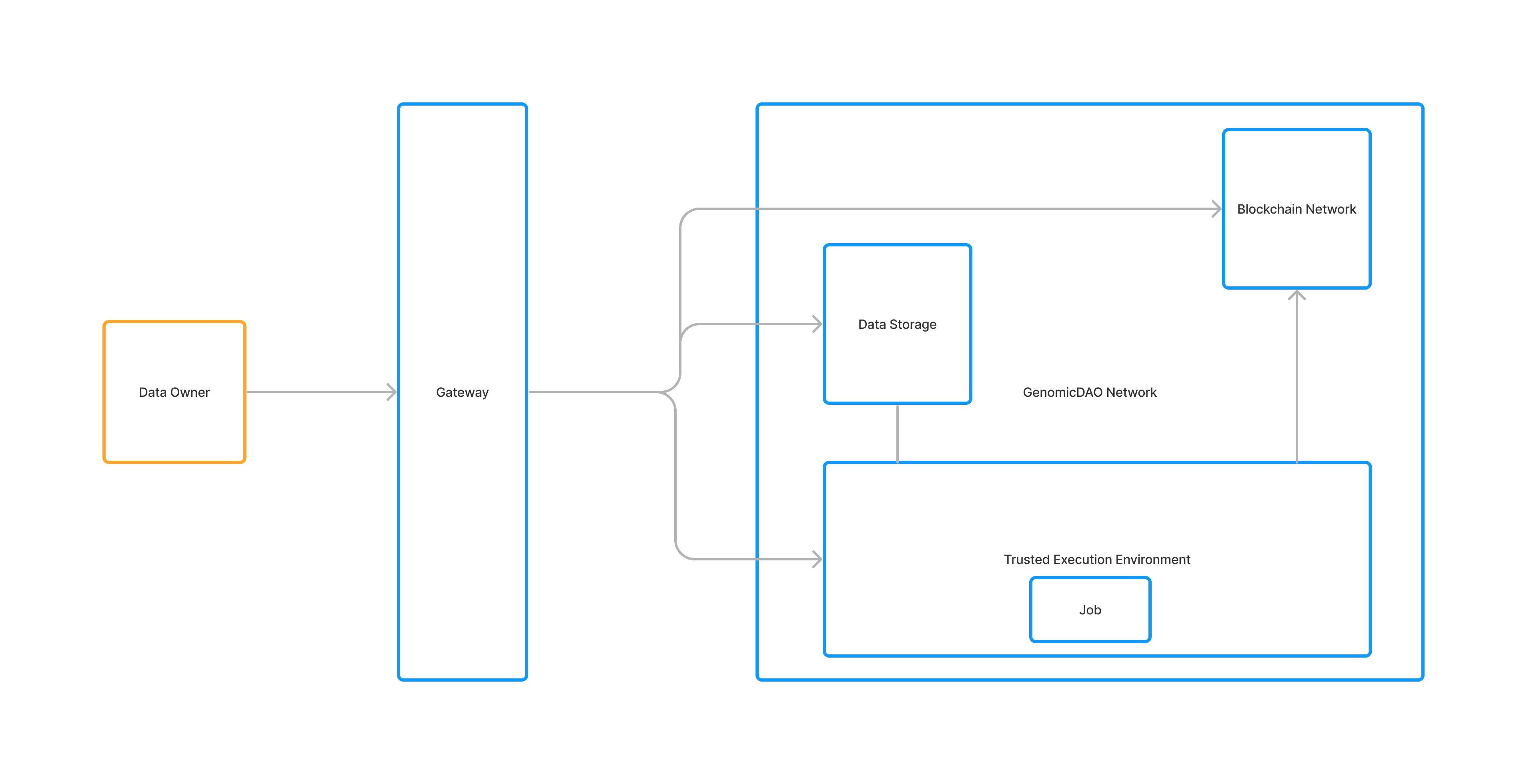
GenomicDAO is an AI-Powered DeSci Platform for Precision Medicine. PCSP (Post-Covid Stroke Prevention), the first released token on GenomicDAO platform, allows users to purchase genetic testing services and to govern the drug discovery and development for stroke. The G-Stroke service, a product of the GenomicDAO, provides the information about the risk of stroke to the user based his/her genetic data.
- After the registered user purchases G-Stroke service, the saliva sample will be collected for genetic decoding. User’s gene data is encrypted and securely stored in the system.
- All the computation on the user’s gene data is executed in a TEE (Trusted execution environment) where the computation cannot be tampered with and the intermediate stages of a computation cannot be read by not-a-computer.
- The result of the computation process is the genetic insights which will be used to generate a full report for the user.
- According to the stroke risk score, the user will be received amount of PCSP token corresponding to the following:
- extremely high risk: 15000 PCSP
- high risk: 3000 PCSP
- slightly high risk: 225 PCSP
- normal or low risk: 30 PCSP
- At the same time, the user receives a G-NFT, which is minted on the blockchain based on the user's gene data. This NFT defines the ownership of the gene profile.
Data format
The gene data, stored as a text file, comprises the user’s genetic information. Importantly, it lacks any identifiers that could link back to the individual’s real-world identity. Each user’s gene data is distinct, ensuring no duplication exists within the system. Typically, the size of this text file is around 40MB. To simplify this assignment, the gene text file will only contain one of the following text content:
- "extremely high risk"
- "high risk"
- "slightly high risk"
- "low risk".
Upon processing this data through the TEE, the program will assign a corresponding risk score based on the gene data provided.
The client makes requests to the Gateway using conventional REST endpoints.
Where the encrypted gene data is stored.
To protect algorithms and data from unauthorized observers, ensuring that inputs and outputs are end-to-end encrypted, we use the trusted execution environment (TEE).
The following main functions need to be implemented:
- Submit gene data
- Mint G-NFT
- Reward PCSP
Those features should be implemented with contracts on layer 2 built with OP Stack. For more information:
- To complete this assignment you should implement the main features of the components to demonstrate the user’s end-to-end flow.
- Propose solutions to protect the user’s gene data so that no one could access it directly without the end user’s permission.
- Implement the solution on the Optimism OP Stack
To complete this assignment you should implement the /// TODO: with solution code. Most of the changes will be in /contracts/
-
Contracts:
- NFT.sol: for GeneNFT
- Token.sol: for PCSP Token
- Storage.sol: for managing the document info on-chain
-
Blockchain event when:
- Gene data is submitted
- GNFT is minted
- PCSP is rewarded
Requirements
Go to folder /genomicdao/ and run the following command to install package requirements
npm install
To run the test
npx hardhat test
Build OP Stack Rollups. For more information Optimism docs
Clone source code
git clone https://github.com/ethereum-optimism/optimism.git
Build packages
cd optimism
pnpm install
make op-node op-batcher op-proposer
pnpm build
Clone source code
cd ~
git clone https://github.com/ethereum-optimism/op-geth.git
Build from source code
cd op-geth
make geth
- The
Adminaccount which has the ability to upgrade contracts. - The
Batcheraccount which publishes Sequencer transaction data to L1. - The
Proposeraccount which publishes L2 transaction results to L1. - The
Sequenceraccount which signs blocks on the p2p network.
Also, fund these accounts some test ETH for running.
cd ~/op-geth
./build/bin/geth \
--datadir ./datadir \
--http \
--http.corsdomain="*" \
--http.vhosts="*" \
--http.addr=0.0.0.0 \
--http.api=web3,debug,eth,txpool,net,engine \
--ws \
--ws.addr=0.0.0.0 \
--ws.port=8546 \
--ws.origins="*" \
--ws.api=debug,eth,txpool,net,engine \
--syncmode=full \
--gcmode=archive \
--nodiscover \
--maxpeers=0 \
--networkid=42069 \
--authrpc.vhosts="*" \
--authrpc.addr=0.0.0.0 \
--authrpc.port=8551 \
--authrpc.jwtsecret=./jwt.txt \
--rollup.disabletxpoolgossip=true
cd ~/optimism/op-node
./bin/op-node \
--l2=http://localhost:8551 \
--l2.jwt-secret=./jwt.txt \
--sequencer.enabled \
--sequencer.l1-confs=3 \
--verifier.l1-confs=3 \
--rollup.config=./rollup.json \
--rpc.addr=0.0.0.0 \
--rpc.port=8547 \
--p2p.disable \
--rpc.enable-admin \
--p2p.sequencer.key=$SEQ_KEY \
--l1=$L1_RPC \
--l1.rpckind=$RPC_KIND
cd ~/optimism/op-batcher
./bin/op-batcher \
--l2-eth-rpc=http://localhost:8545 \
--rollup-rpc=http://localhost:8547 \
--poll-interval=1s \
--sub-safety-margin=6 \
--num-confirmations=1 \
--safe-abort-nonce-too-low-count=3 \
--resubmission-timeout=30s \
--rpc.addr=0.0.0.0 \
--rpc.port=8548 \
--rpc.enable-admin \
--max-channel-duration=1 \
--l1-eth-rpc=$L1_RPC \
--private-key=$BATCHER_KEY
cd ~/optimism/op-proposer
./bin/op-proposer \
--poll-interval=12s \
--rpc.port=8560 \
--rollup-rpc=http://localhost:8547 \
--l2oo-address=$L2OO_ADDR \
--private-key=$PROPOSER_KEY \
--l1-eth-rpc=$L1_RPC
genomicdao/ # Directory contains main
│
├── contracts/
│ ├── NFT.sol
│ ├── Token.sol
│ ├── Storage.sol
│
├── scripts
│ ├── deploy.js
└── ...