The official repository of PheTK.
Quick links:
- Installation
- System requirements
- 1-minute PheWAS demo
- PheTK description
- Usage examples
- Changelog (refer to this section for the latest version of PheTK)
version 0.1.45 (01 Oct 2024):
- Fixed a minor issue where Bonferroni line is shifted in Manhattan Plot after non-converged phecodes are removed.
- Updated README (this section) to add description for other plotting features, such as setting custom Bonferroni value or custom color palette.
version 0.1.44 (30 Sep 2024):
- Removed polars version requirement of <=0.20.26 since polars has fixed multithreading bug.
- Updated Demo with more descriptive texts.
- Updated Plot module (see this section for usage examples):
- Added parameter
converged_onlythat can be used when instantiatePlotclass to plot only converged phecodes. - Added parameter
marker_size_by_betaandmarker_scale_factorinmanhattan()function
- Added parameter
- Updated README to reflect changes.
version 0.1.43 (08 Aug 2024) - IMPORTANT BUG FIX:
-
Fixed an issue in
.by_genotype()in Cohort module which might generate incorrect cohort by genotype for multi-allelic sites from All of Us variant data or custom Hail matrix table. For example, for a site that has 3 alleles ["A", "G", "C"] (reference allele, alt allele 1, alt allele 2, as displayed in Hail), if user specifies "A" asref_allele, "C" asalt_allele, and "0/1" ascase_gt:- Before this fix, given above user inputs, participants with A-C genotype would not be assigned as cases, since the allele index of "C" is still "2", and therefore A-C genotype would be encoded as "0/2", even after multi-allelic split.
- After this fix, given above user inputs, participants with A-C genotype will be correctly assigned as cases, since the allele index for "C" would be properly updated to "1" after multi-allelic split.
-
This issue affects users who used method
.by_genotype()to generate cohort:- from All of Us data, having ALL the criteria below:
- the genomic position was a multi-allelic site,
- the alternative allele of interest was NOT the first alternative allele ("G" in the above example).
- from custom unsplit matrix table or improperly split matrix table as input, having the same above criteria.
- from All of Us data, having ALL the criteria below:
-
Going forward, there is nothing changed in how user would use
.by_genotype(), i.e., "0" represents reference allele, and "1" represents alternative allele of interest. -
Users should uninstall any previous version, reinstall PheTK, and make sure current version is v0.1.43. It is recommended for affected users to rerun
.by_genotype()step and potentially subsequent steps.
version 0.1.42 (17 Jul 2024):
- Added method
.get_phecode_data()in PheWAS class. This method would generate cohort data from input data for a phecode of interest. Please refer to this section in PheWAS module for usage example.
version 0.1.41 (22 May 2024):
- Added polars version <= 0.20.26 as dependency requirement since polars version 0.20.27 could cause multithreading issue.
version 0.1.40 (14 May 2024):
- Fixed an incorrect printout text check in
.by_genotype()in Cohort module when a variant is not found. This is only an aesthetic fix to avoid confusion, and does not affect previously generated cohorts.
version 0.1.39 (02 May 2024):
- Added lxml as a required dependency during installation as it might not be preinstalled in some platforms.
version 0.1.38 (11 Apr 2024):
- Updated default All of Us common variant matrix table (ACAF) file path used by
.by_genotype()method in Cohort module with All of Us updated path. - Updated README to add some minimum VM configuration suggestions.
version 0.1.37 (04 Apr 2024):
- Removed SNOMED codes from SQL query (ehr_dx_code_query) generating ehr_length, dx_code_occurrence_count,
dx_condition_count, and age_at_last_event covariates in
.add_covariates()method in Cohort module.- This was to make it consistent with ICD event query for phecode mapping, i.e., only ICD9CM and ICD10CM would be used as vocabulary_id for these queries.
- For All of Us users, this change should affect less than 2% of covariate data previously generated
by
.add_covariates()method from version 0.1.36 or earlier, and should not significantly change previous analysis results.
PheTK can be installed using pip install command in a terminal (Python 3.7 or newer):
pip install PheTK
or Jupyter Notebook (restart kernel after installation and prior to importing):
!pip install PheTK
If and older version of PheTK was installed previously, it is best to uninstall it before new installation
pip uninstall PheTK -y && pip install PheTK
To check current installed version:
pip show PheTK | grep Version
PheTK was developed for efficient processing of large data while being resource friendly. It was tested on different platforms from laptops to different cloud environments.
Here are some minimum VM configuration suggestions for All of Us users:
- Cohort module: default General Analysis (4 CPUs, 15GB RAM) or Hail Genomic Analysis (main VM and 2 workers of 4 CPUs, 15GB RAM each) VMs should work. Hail Genomic Analysis is only needed for .by_genotype() method.
- Phecode module: a minimum of 8 CPUs, 52GB RAM standard VM should work for current v7 data. This might require more RAM depending on user custom workflow or data - usually Jupyter Python kernel would die at if there is not enough memory.
- PheWAS module: default General Analysis VM (4 CPUs, 15GB RAM) should work. However, more CPUs would speed up analysis and using low configurations do not necessarily save computing cost since total runtime would be longer.
- Plot module: default General Analysis VM (4 CPUs, 15GB RAM) should work.
In practice, users could try different available machine configurations to achieve optimal performance and cost-effectiveness.
User can run the quick 1-minute PheWAS demo with the following command in a terminal:
python3 -m PheTK.Demo
Or in Jupyter Notebook:
from PheTK import Demo
Demo.run()
The example files (example_cohort.csv, example_phecode_counts.csv, and example_phewas_results.csv)
generated in this Demo should be in users' current working directory.
New-to-PheWAS users could explore these files to get a sense of what data are used or generated in PheWAS with PheTK.
PheTK is a fast python library for Phenome Wide Association Studies (PheWAS) utilizing both phecode 1.2 and phecodeX 1.0.
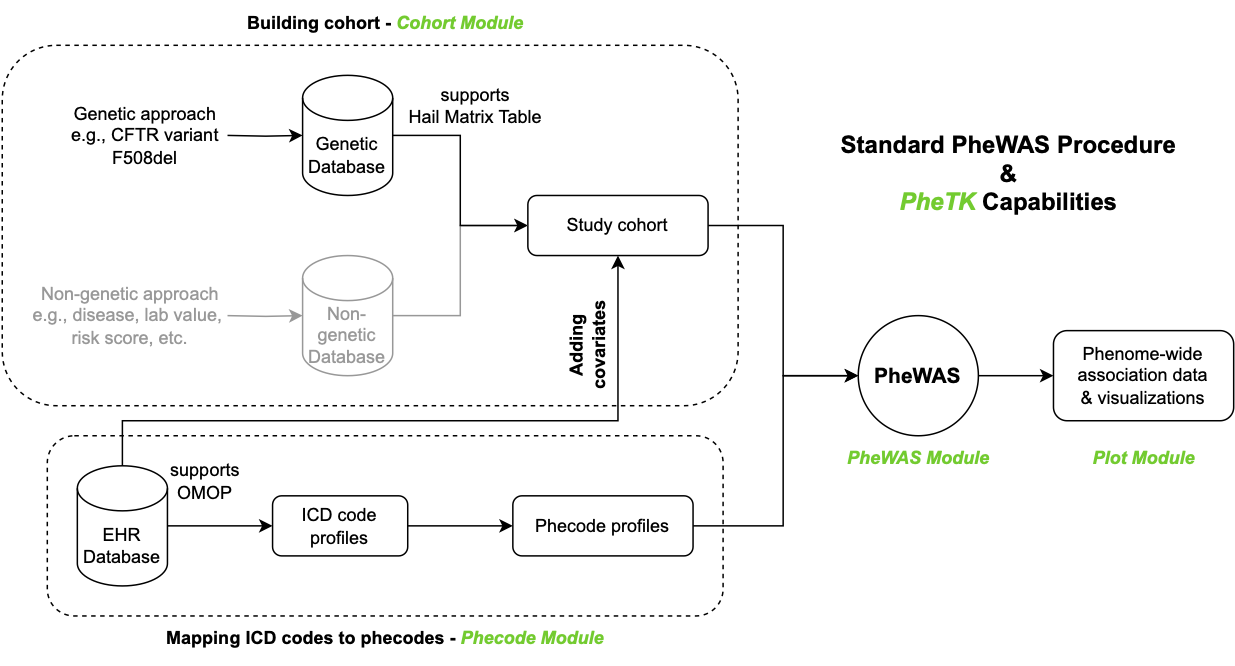 Standard PheWAS workflow. Green texts are PheTK module names.
Black components are supported while gray ones are not supported by PheTK currently.
Standard PheWAS workflow. Green texts are PheTK module names.
Black components are supported while gray ones are not supported by PheTK currently.
This table will be updated as we update PheTK.
All modules can be used together or independently, e.g., users who only need to run PheWAS analysis can provide their own cohort and phecode count data as input for PheWAS module.
| Module | Class | Method(s) | Platform | Requirements/Notes |
|---|---|---|---|---|
| Cohort | Cohort | by_genotype | All of Us | None |
| Other | Variant data stored in Hail matrix table | |||
| add_covariate | All of Us | None | ||
| Other | Google BigQuery OMOP database | |||
| Required tables: person, condition_occurrence, observation, death, & concept | ||||
| Phecode | Phecode | count_phecode | All of Us | None |
| Other | User provided cohort ICD code data | |||
| User can use custom ICD-to-phecode mapping table. | ||||
| PheWAS | PheWAS | all methods | Any | None |
| Plot | Plot | all methods | Any | None |
| Demo | all methods | Any | None |
All of Us: the All of Us Research Program (https://allofus.nih.gov/)
Cohort module can be used for generating genetic cohort and add certain covariates to a cohort.
This function takes genetic variant information as input, and generates cohort with matching genotypes as an output csv file. As this function uses Hail to extract data from Hail matrix tables, it must be run in a compatible environment, e.g., a dataproc cluster on All of Us researcher workbench or UK Biobank RAP.
For example, we generate cohort for CFTR variant chr7-117559590-ATCT-A with heterozygous (0/1 genotype) participants as cases and homozygous reference (0/0 genotype) participants as controls.
For All of Us data version 7, the default Hail matrix table is the ACAF (common variant) table. User can use a different table by providing table location in the mt_path parameter.
from PheTK.Cohort import Cohort
# instantiate class Cohort object for _All of Us_ database version 7
cohort = Cohort(platform="aou", aou_db_version=7)
# generate cohort by genotype
cohort.by_genotype(
chromosome_number=7,
genomic_position=117559590,
ref_allele="ATCT",
alt_allele="A",
case_gt="0/1",
control_gt="0/0",
reference_genome="GRCh38",
mt_path=None,
output_file_name="cftr_cohort.csv"
)
For other platforms, user need to provide the location of Hail matrix table file for mt_path parameter.
from PheTK.Cohort import Cohort
# instantiate class Cohort object for _All of Us_ database version 7
cohort = Cohort(platform="custom")
# generate cohort by genotype
cohort.by_genotype(
chromosome_number=7,
genomic_position=117559590,
ref_allele="ATCT",
alt_allele="A",
case_gt="0/1",
control_gt="0/0",
reference_genome="GRCh38",
mt_path="/path/to/hail_matrix_table.mt",
output_file_name="cftr_cohort.csv"
)
This function is currently customized for the All of Us Research Platform. It takes a cohort csv file and covariate selection as input, and generate a new cohort csv file with covariate data added as output. Input cohort data must have "person_id" column.
For non-All of Us platforms, a Google BigQuery dataset ID must be provided.
In this example, we are adding age at last diagnosis event, sex at birth and 10 genetic PCs (provided by All of Us). These options were set to True (or 10 in case of first_n_pcs).
The covariates shown in this example are currently supported by PheTK. Users should only change parameter value to True for covariates to be used in subsequent PheWAS. All parameters are set to False by default, i.e., user only need to specify parameters of interest as shown in the "short version".
It is highly recommended that users should decide which covariates to use for the study based on their data, and it is perfectly fine to add or use their own covariate data if necessary.
# user can skip the import and instantiation steps if running continuously
# from previous by_genotype example, i.e., skip directly to add covariates step.
from PheTK.Cohort import Cohort
# instantiate class Cohort object for _All of Us_ database version 7
cohort = Cohort(platform="aou", aou_db_version=7)
# RUN EITHER LONG OR SHORT VERSION BELOW
# add covariates - long version, including all currently supported covariate options
cohort.add_covariates(
cohort_csv_path="cftr_cohort.csv",
natural_age=False,
age_at_last_event=True,
sex_at_birth=True,
ehr_length=False,
dx_code_occurrence_count=False,
dx_condition_count=False,
genetic_ancestry=False,
first_n_pcs=10,
drop_nulls=True,
output_file_name="cohort_with_covariates.csv"
)
# add covariates - short version, i.e., users do not need to list unused covariates
cohort.add_covariates(
cohort_csv_path="cftr_cohort.csv",
age_at_last_event=True,
sex_at_birth=True,
first_n_pcs=10,
drop_nulls=True,
output_file_name="cohort_with_covariates.csv"
)
Covariate descriptions:
- natural_age: current age or age at death
- age_at_last_event: age at last diagnosis event (ICD or SNOMED) in EHR.
- sex_at_birth: sex at birth
- ehr_length: EHR duration, in year, from first to last diagnosis code
- dx_code_occurrence_count: counts the occurrences of diagnosis codes throughout EHR of each participant. For example: person 1 having R50 (fever) code on 5 different dates, R05 (cough) code on 3 different dates, and R05.1 (acute cough) code on 2 different dates, will have a dx_code_occurrence_count = 10.
- dx_condition_count: counts the number of unique conditions occurred throughout EHR of each participant. For example, for the same person 1 above, the dx_condition_count = 3 (R05 - cough, R05.1 - acute cough, R50 - fever).
- genetic_ancestry: returns string values of predicted ancestries, e.g., "eur", "afr", etc. These are only useful if user would like to filter data by genetic ancestries.
- first_n_pcs: retrieves first n genetic PC components from genetic PCA data generated by All of Us.
- drop_nulls: remove rows containing null values in any column.
The only difference in this case is that user need to provide dataset ID for gbq_dataset_id parameter. The rest should be the same as above example. Please make sure the custom database meet the requirements in section 4.2.
from PheTK.Cohort import Cohort
cohort = Cohort(platform="custom", gbq_dataset_id="Google_BigQuery_dataset_id")
Phecode module is used to retrieve ICD code data of participants, map ICD codes to phecode 1.2 or phecodeX 1.0, and aggregate the counts for each phecode of each participant.
The ICD code retrieval is done automatically for All of Us platform when users instantiate class Phecode. For other platforms, users must provide your own ICD code data.
Example of ICD code data:
- Each row must be unique, i.e., there should not be 2 instances of 1 ICD code in the same day.
- Data must have these exact column names.
- "vocabulary_id" column can be replaced with "flag" column with values of 9 and 10.
Example with "vocabulary_id" column:
| person_id | date | vocabulary_id | ICD |
|---|---|---|---|
| 13579 | 1-11-2010 | ICD9CM | 786.2 |
| 13579 | 1-31-2010 | ICD9CM | 786.2 |
| 13579 | 12-4-2017 | ICD10CM | R05.1 |
| 24680 | 3-12-2012 | ICD9CM | 659.2 |
| 24680 | 4-18-2018 | ICD10CM | R50 |
Example with "flag" column:
| person_id | date | flag | ICD |
|---|---|---|---|
| 13579 | 1-11-2010 | 9 | 786.2 |
| 13579 | 1-31-2010 | 9 | 786.2 |
| 13579 | 12-4-2017 | 10 | R05.1 |
| 24680 | 3-12-2012 | 9 | 659.2 |
| 24680 | 4-18-2018 | 10 | R50 |
In these examples, we will map US ICD codes (ICD-9-CM & ICD-10-CM) to phecodeX for All of Us and custom platforms.
from PheTK.Phecode import Phecode
phecode = Phecode(platform="aou")
phecode.count_phecode(
phecode_version="X",
icd_version="US",
phecode_map_file_path=None,
output_file_name="my_phecode_counts.csv"
)
from PheTK.Phecode import Phecode
phecode = Phecode(platform="custom", icd_df_path="/path/to/my_icd_data.csv")
phecode.count_phecode(
phecode_version="X",
icd_version="US",
phecode_map_file_path=None,
output_file_name="my_phecode_counts.csv"
)
Users can provide their own phecode mapping file by adding a csv file path to phecode_map_file_path.
If user provides their own ICD data, platform should be set to "custom".
Custom phecode mapping file must have columns:
phecode: contains code values of phecodesICD: contains code values of ICD codesflag: specifies whether ICD codes is of version 9 or 10; takes numerical 9 or 10 as values.sex: sex of ICD/phecode; takes "Male", "Female", or "Both" as values.phecode_string: literal name of phecodes as stringsphecode_category: specifies which category a phecode belongs to.exclude_range: only applicable for phecode 1.2.; specifies the range of exclusion if used in PheWAS; for example, "008.5,008.7,008.51,008.52,008.6"
It is recommended to run Demo example above and have a look at example cohort and phecode counts file to be familiar with input data format. The example files should be generated in user's current working directory.
PheWAS class is instantiated with paths to csv files of cohort data and phecode counts data, in addition to other parameters as shown in the examples below. It can be used in both Linux command line interface (CLI) and any Python environment, e.g., Jupyter Notebook/Lab.
In these example, we would like to run PheWAS with phecodeX for example data generated by Demo module, with age, sex and 3 genetic PCs as covariates, and an independent variable of interest (for which PheWAS summary statistics will be generated). For each phecode, a participant must have a minimum count of 2 phecode events to be considered a case. There must be at least 50 cases and 50 controls for the phecode to be tested.
python3 -m PheTK.PheWAS \
--phecode_version X \
--cohort_csv_path example_cohort.csv \
--phecode_count_csv_path example_phecode_counts.csv \
--sex_at_birth_col sex \
--male_as_one True \
--covariates age sex pc1 pc2 pc3 \
--independent_variable_of_interest independent_variable_of_interest \
--min_case 50 \
--min_phecode_count 2 \
--output_file_name example_phewas_results.csv
from PheTK.PheWAS import PheWAS
example_phewas = PheWAS(
phecode_version="X",
phecode_count_csv_path="example_phecode_counts.csv",
cohort_csv_path="example_cohort.csv",
sex_at_birth_col="sex",
male_as_one=True,
covariate_cols=["age", "sex", "pc1", "pc2", "pc3"],
independent_variable_of_interest="independent_variable_of_interest",
min_cases=50,
min_phecode_count=2,
output_file_name="example_phewas_results.csv"
)
example_phewas.run()
Notes:
- Each entry in sex_at_birth column should be either 0 or 1 for female or male. The default is male = 1 and female = 0.
- User can use male_as_one to specify where male was coded as 1 (male_as_one=True) or 0 (male_as_one=False).
- In the above example, "sex" column was declared twice, once in sex_at_birth_col and once in covariate_cols. sex_at_birth_col is always required as certain phecodes are sex restricted. If user would like to use sex as a covariate, sex column must be included in covariate_cols.
- PheWAS results often include both converged and non-converged phecodes.
This was intentional, as there can be multiple factors affect regression model convergence.
Therefore, non_converged phecodes are kept and flagged using
convergedcolumn to allow users to do further investigation if needed.
Method .get_phecode_data() can be used to get cohort data for a phecode of interest.
A common use case is to look at input data of certain phecodes after a PheWAS run.
In addition to the existing columns, e.g., person_id, covariates, etc., from input data,
the generated table has is_phecode_case boolean column to specify whether an individual is a case or control for that phecode.
For example, after creating a PheWAS class instance as in the above example,
user can call .get_phecode_data() method as below:
# get cohort data for phecode "R05.1"
example_phewas.get_phecode_data("R05.1")
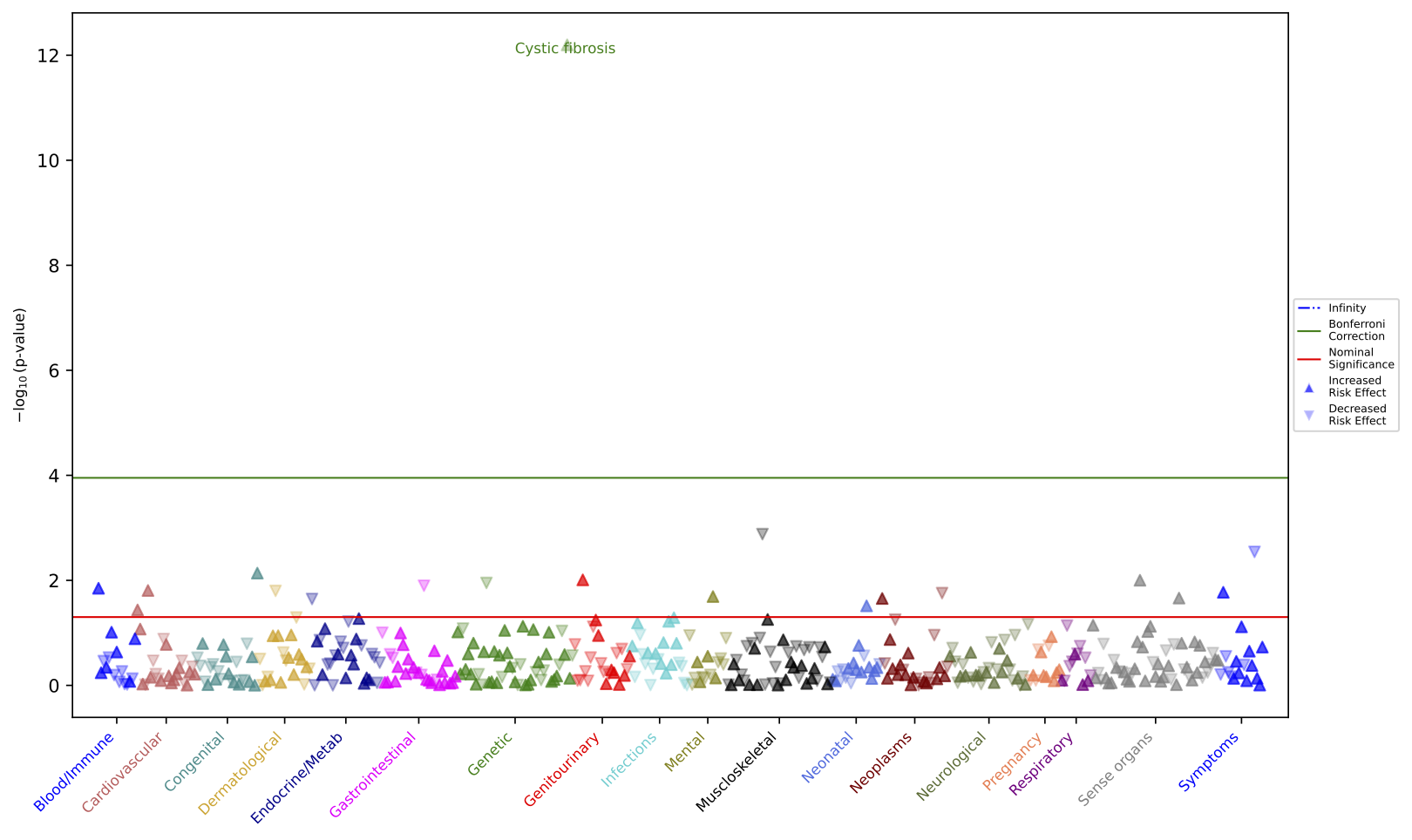
Plot class is instantiated with path to PheWAS result csv file. After that, a plot type method can be called to generate a plot, e.g., calling manhattan() method to make Manhattan plot.
In this example, we are generating a Manhattan plot for the PheWAS results created by module Demo.
from PheTK.Plot import Plot
p = Plot("example_phewas_results.csv")
p.manhattan(label_values="p_value", label_count=1, save_plot=True)
The above code example generates this Manhattan plot figure:
The features below can be used individually or in combinations to customize Manhattan Plot.
As mentioned in PheWAS module above, PheWAS results contain both converged and non-converged phecodes. By default, PheTK will exclude non-converged phecodes when plotting for better interpretation.
Users can include non-converged phecodes by setting boolean parameter converged_only to False:
p = Plot("example_phewas_results.csv", converged_only=False)
p = Plot("example_phewas_results.csv", bonferroni=your_custom_value)
Color palette should be provided in a tuple format. Each color will be used for a phecode category. If the number of colors is less than number of phecode categories, the colors will be cycled back to the first color and so on. PheTK uses matplotlib color names.
p = Plot("example_phewas_results.csv", color_palette=("blue", "indianred", "darkcyan"))
This feature can be turned on using boolean parameter marker_size_by_beta to True.
Users can adjust marker size using parameter marker_scale_factor which has a default value of 1.
(Note that this parameter only works when marker_size_by_beta=True).
p.manhattan(marker_size_by_beta=True, marker_scale_factor=1)
By default, PheTK will label by top p-values. This can be changed using parameter label_values.
This parameter accepts single string value or list of phecodes, or 3 preset values of "p_value", "positive_beta", or "negative_beta".
Note that, if provided text values do not match the preset values or phecodes in PheWAS results, they will not be labeled.
Label by top p-values:
p.manhattan(label_values="p_value")
Label by top positive beta values:
p.manhattan(label_values="positive_beta")
Label by top negative beta values:
p.manhattan(label_values="negative_beta")
Label by specific phecode(s) of interests:
p.manhattan(label_values=["GE_982", "NS_351"])
By default, PheTK will label top 10 data points of label values. To change this, use parameter label_count
p.manhattan(label_count=10)
Users can choose to label only in a specific phecode category or multiple categories using parameter phecode_categories
p.manhattan(phecode_categories=["Neoplasms", "Genetic"])
p.manhattan(save_plot=True)
Further details on plot customization options will be provided in a separate document in the near future.
Preprint version: https://www.medrxiv.org/content/10.1101/2024.02.12.24302720v1
Peer reviewed version: TBD