phylocomr
phylocomr gives you access to the Phylocom C library.
Package API
ecovolve/ph_ecovolve- interface toecovolveexecutable, and a higher level interfacephylomatic/ph_phylomatic- interface tophylomaticexecutable, and a higher level interfacephylocom- interface tophylocomexecutableph_aot- higher level interface toaotph_bladj- higher level interface tobladjph_comdist/ph_comdistnt- higher level interface to comdistph_comstruct- higher level interface to comstructph_comtrait- higher level interface to comtraitph_pd- higher level interface to Faith's phylogenetic diversity
Installation
Stable version:
install.packages("phylocomr")Development version:
remotes::install_github("ropensci/phylocomr")library("phylocomr")
library("ape")ecovolve
ph_ecovolve(speciation = 0.05, extinction = 0.005, time_units = 50)#> $phylogeny
#> [1] "((dead2:4.000000,sp7:19.000000)node1:15.000000,(dead5:5.000000,(sp8:11.000000,(sp9:3.000000,sp10:3.000000)node6:8.000000)node4:3.000000)node3:20.000000)node0:16.000000;\n"
#>
#> $sample
#> # A tibble: 4 x 3
#> sample abundance name
#> <chr> <int> <chr>
#> 1 alive 1 sp7
#> 2 alive 1 sp8
#> 3 alive 1 sp9
#> 4 alive 1 sp10
#>
#> $traits
#> # A tibble: 6 x 6
#> name ecov0 ecov1 ecov2 ecov3 ecov4
#> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 dead2 2 -17 5 17 -8
#> 2 dead5 -4 -3 -5 1 3
#> 3 sp7 6 -14 18 16 -15
#> 4 sp8 10 -21 4 -6 -1
#> 5 sp9 6 -18 5 -4 0
#> 6 sp10 -1 -23 8 -4 -1
phylomatic
taxa_file <- system.file("examples/taxa", package = "phylocomr")
phylo_file <- system.file("examples/phylo", package = "phylocomr")
(taxa_str <- readLines(taxa_file))#> [1] "campanulaceae/lobelia/lobelia_conferta"
#> [2] "cyperaceae/mapania/mapania_africana"
#> [3] "amaryllidaceae/narcissus/narcissus_cuatrecasasii"
(phylo_str <- readLines(phylo_file))#> [1] "(((((eliea_articulata,homalanthus_populneus)malpighiales,rosa_willmottiae),((macrocentrum_neblinae,qualea_clavata),hibiscus_pohlii)malvids),(((lobelia_conferta,((millotia_depauperata,(layia_chrysanthemoides,layia_pentachaeta)layia),senecio_flanaganii)asteraceae)asterales,schwenkia_americana),tapinanthus_buntingii)),(narcissus_cuatrecasasii,mapania_africana))poales_to_asterales;"
ph_phylomatic(taxa = taxa_str, phylo = phylo_str)#> [1] "(lobelia_conferta:5.000000,(mapania_africana:1.000000,narcissus_cuatrecasasii:1.000000):1.000000)poales_to_asterales;\n"
#> attr(,"taxa_file")
#> [1] "/var/folders/fc/n7g_vrvn0sx_st0p8lxb3ts40000gn/T//Rtmp58xZbT/taxa_20f86538e35e"
#> attr(,"phylo_file")
#> [1] "/var/folders/fc/n7g_vrvn0sx_st0p8lxb3ts40000gn/T//Rtmp58xZbT/phylo_20f879d7643c"
aot
traits_file <- system.file("examples/traits_aot", package = "phylocomr")
phylo_file <- system.file("examples/phylo_aot", package = "phylocomr")
traitsdf_file <- system.file("examples/traits_aot_df", package = "phylocomr")
traits <- read.table(text = readLines(traitsdf_file), header = TRUE,
stringsAsFactors = FALSE)
phylo_str <- readLines(phylo_file)
ph_aot(traits = traits, phylo = phylo_str)#> $trait_conservatism
#> # A tibble: 124 x 28
#> trait trait.name node name age ntaxa n.nodes tip.mn tmn.ranklow
#> <int> <chr> <int> <chr> <dbl> <int> <int> <dbl> <int>
#> 1 1 traitA 0 A 5 32 2 1.75 1000
#> 2 1 traitA 1 B 4 16 2 1.75 640
#> 3 1 traitA 2 C 3 8 2 1.75 678
#> 4 1 traitA 3 D 2 4 2 1.5 266
#> 5 1 traitA 4 E 1 2 2 1 61
#> 6 1 traitA 7 F 1 2 2 2 1000
#> 7 1 traitA 10 G 2 4 2 2 1000
#> 8 1 traitA 11 H 1 2 2 2 1000
#> 9 1 traitA 14 I 1 2 2 2 1000
#> 10 1 traitA 17 J 3 8 2 1.75 651
#> # ... with 114 more rows, and 19 more variables: tmn.rankhi <int>,
#> # tip.sd <dbl>, tsd.ranklow <int>, tsd.rankhi <int>, node.mn <dbl>,
#> # nmn.ranklow <int>, nmn.rankhi <int>, nod.sd <dbl>, nsd.ranklow <int>,
#> # nsd.rankhi <int>, sstipsroot <dbl>, sstips <dbl>,
#> # percvaramongnodes <dbl>, percvaratnode <dbl>, contributionindex <dbl>,
#> # sstipvnoderoot <dbl>, sstipvnode <dbl>, ssamongnodes <dbl>,
#> # sswithinnodes <dbl>
#>
#> $independent_contrasts
#> # A tibble: 31 x 17
#> node name age n.nodes contrast1 contrast2 contrast3 contrast4
#> <int> <chr> <dbl> <int> <dbl> <dbl> <dbl> <dbl>
#> 1 0 A 5 2 0 0 0 0.254
#> 2 1 B 4 2 0 1.03 0 0.516
#> 3 2 C 3 2 0.267 0.535 0 0
#> 4 3 D 2 2 0.577 0 1.15 0
#> 5 4 E 1 2 0 0 0.707 0
#> 6 7 F 1 2 0 0 0.707 0
#> 7 10 G 2 2 0 0 1.15 0
#> 8 11 H 1 2 0 0 0.707 0
#> 9 14 I 1 2 0 0 0.707 0
#> 10 17 J 3 2 0.267 0.535 0 0
#> # ... with 21 more rows, and 9 more variables: contrastsd <dbl>,
#> # lowval1 <dbl>, hival1 <dbl>, lowval2 <dbl>, hival2 <dbl>,
#> # lowval3 <dbl>, hival3 <dbl>, lowval4 <dbl>, hival4 <dbl>
#>
#> $phylogenetic_signal
#> # A tibble: 4 x 5
#> trait ntaxa varcontr varcn.ranklow varcn.rankhi
#> <chr> <int> <dbl> <int> <int>
#> 1 traitA 32 0.054 2 999
#> 2 traitB 32 0.109 1 1000
#> 3 traitC 32 0.622 68 933
#> 4 traitD 32 0.011 1 1000
#>
#> $ind_contrast_corr
#> # A tibble: 3 x 6
#> xtrait ytrait ntaxa picr npos ncont
#> <chr> <chr> <int> <dbl> <dbl> <int>
#> 1 traitA traitB 32 0.248 18.5 31
#> 2 traitA traitC 32 0.485 27.5 31
#> 3 traitA traitD 32 0 16.5 31
bladj
ages_file <- system.file("examples/ages", package = "phylocomr")
phylo_file <- system.file("examples/phylo_bladj", package = "phylocomr")
ages_df <- data.frame(
a = c('malpighiales','salicaceae','fabaceae','rosales','oleaceae',
'gentianales','apocynaceae','rubiaceae'),
b = c(81,20,56,76,47,71,18,56)
)
phylo_str <- readLines(phylo_file)
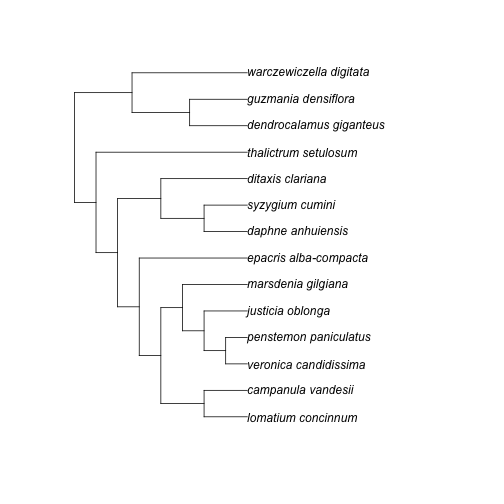
(res <- ph_bladj(ages = ages_df, phylo = phylo_str))#> [1] "((((((lomatium_concinnum:20.250000,campanula_vandesii:20.250000):20.250000,(((veronica_candidissima:10.125000,penstemon_paniculatus:10.125000)plantaginaceae:10.125000,justicia_oblonga:20.250000):10.125000,marsdenia_gilgiana:30.375000):10.125000):10.125000,epacris_alba-compacta:50.625000)ericales_to_asterales:10.125000,((daphne_anhuiensis:20.250000,syzygium_cumini:20.250000)malvids:20.250000,ditaxis_clariana:40.500000):20.250000):10.125000,thalictrum_setulosum:70.875000)eudicots:10.125000,((dendrocalamus_giganteus:27.000000,guzmania_densiflora:27.000000)poales:27.000000,warczewiczella_digitata:54.000000):27.000000)malpighiales:1.000000;\n"
#> attr(,"ages_file")
#> [1] "/var/folders/fc/n7g_vrvn0sx_st0p8lxb3ts40000gn/T//Rtmp58xZbT/ages"
#> attr(,"phylo_file")
#> [1] "/var/folders/fc/n7g_vrvn0sx_st0p8lxb3ts40000gn/T//Rtmp58xZbT/phylo_20f8386d8ada"
plot(ape::read.tree(text = res))Meta
- Please report any issues or bugs.
- License: MIT
- Get citation information for
phylocomrin R doingcitation(package = 'phylocomr') - Please note that this project is released with a Contributor Code of Conduct. By participating in this project you agree to abide by its terms.