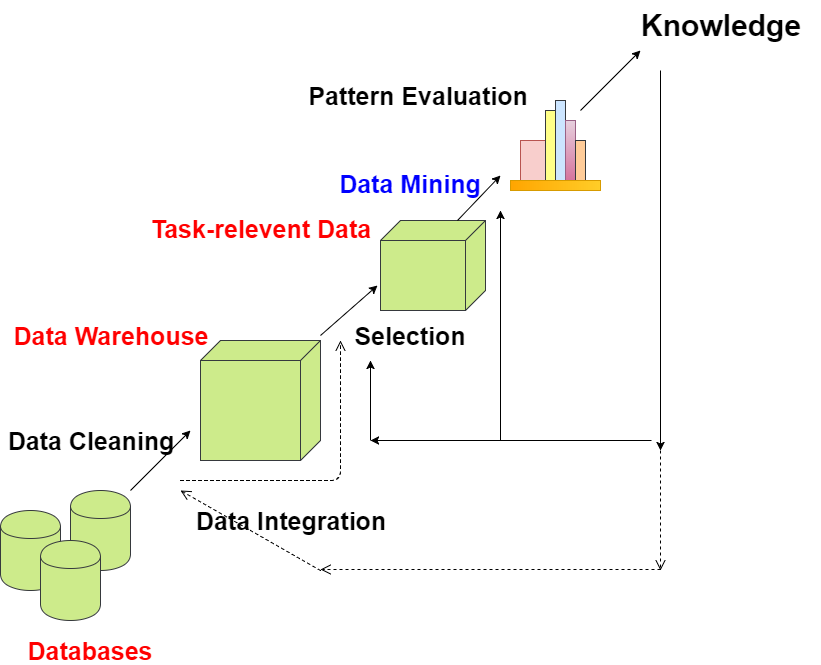
Data mining is the core part of the knowledge discovery process.
KDP is a process of finding knowledge in data, it does this by using data mining methods (algorithms) in order to extract demanding knowledge from large amount of data.
-
Data cleaning - First step in the Knowledge Discovery Process is Data cleaning in which noise and inconsistent data is removed.
-
Data Integration - Second step is Data Integration in which multiple data sources are combined.
-
Data Selection - Next step is Data Selection in which data relevant to the analysis task are retrieved from the database.
-
Data Transformation - In Data Transformation, data are transformed into forms appropriate for mining by performing summary or aggregation operations.
-
Data Mining - In Data Mining, data mining methods (algorithms) are applied in order to extract data patterns.
-
Pattern Evaluation - In Pattern Evaluation, data patterns are identified based on some interesting measures.
-
Knowledge Presentation - In Knowledge Presentation, knowledge is represented to user using many knowledge representation techniques.
# Goal
- Which country is using what vaccine? (World)
- Try to predict the spread of COVID-19 ahead of time to take preventive measures (Italy subset)
## dpc-covid19-ita-andamento-nazionale
| name | type | num_values | distinct_count | total_count | missing_count | int_count | min | max |
|---|---|---|---|---|---|---|---|---|
| data | date | 0 | 695 | 695 | 0 | 0 | 1.5825636E12 | 1.6425216E12 |
| stato | nominal | 1 | 1 | 695 | 0 | 695 | 0.0 | 0.0 |
| ricoverati_con_sintomi | numeric | 0 | 678 | 695 | 0 | 695 | 101.0 | 34697.0 |
| terapia_intensiva | numeric | 0 | 599 | 695 | 0 | 695 | 26.0 | 4068.0 |
| totale_ospedalizzati | numeric | 0 | 674 | 695 | 0 | 695 | 127.0 | 38507.0 |
| isolamento_domiciliare | numeric | 0 | 693 | 695 | 0 | 695 | 94.0 | 2540993.0 |
| totale_positivi | numeric | 0 | 693 | 695 | 0 | 695 | 221.0 | 2562156.0 |
| variazione_totale_positivi | numeric | 0 | 680 | 695 | 0 | 695 | -51884.0 | 172462.0 |
| nuovi_positivi | numeric | 0 | 679 | 695 | 0 | 695 | 78.0 | 220532.0 |
| dimessi_guariti | numeric | 0 | 694 | 695 | 0 | 695 | 1.0 | 6314444.0 |
| deceduti | numeric | 0 | 695 | 695 | 0 | 695 | 7.0 | 141825.0 |
| casi_da_sospetto_diagnostico | numeric | 0 | 160 | 695 | 533 | 162 | 0.0 | 988470.0 |
| casi_da_screening | numeric | 0 | 162 | 695 | 533 | 162 | 0.0 | 653140.0 |
| totale_casi | numeric | 0 | 695 | 695 | 0 | 695 | 229.0 | 9018425.0 |
| tamponi | numeric | 0 | 695 | 695 | 0 | 695 | 4324.0 | 1.57819844E8 |
| casi_testati | numeric | 0 | 640 | 695 | 55 | 640 | 935310.0 | 4.4547215E7 |
| note | nominal | 44 | 44 | 695 | 651 | 44 | 0.0 | 0.0 |
| ingressi_terapia_intensiva | numeric | 0 | 192 | 695 | 283 | 412 | 2.0 | 324.0 |
| note_test | string | 0 | 0 | 695 | 695 | 0 | 0.0 | 0.0 |
| note_casi | string | 0 | 0 | 695 | 695 | 0 | 0.0 | 0.0 |
| totale_positivi_test_molecolare | numeric | 0 | 369 | 695 | 326 | 369 | 2351466.0 | 6786905.0 |
| totale_positivi_test_antigenico_rapido | numeric | 0 | 369 | 695 | 326 | 369 | 957.0 | 2231520.0 |
| tamponi_test_molecolare | numeric | 0 | 369 | 695 | 326 | 369 | 2.8617351E7 | 7.8396506E7 |
| tamponi_test_antigenico_rapido | numeric | 0 | 369 | 695 | 326 | 369 | 116859.0 | 7.9423338E7 |
### Sample
head ../data/dpc-covid19-ita-andamento-nazionale.csv
data,stato,ricoverati_con_sintomi,terapia_intensiva,totale_ospedalizzati,isolamento_domiciliare,totale_positivi,variazione_totale_positivi,nuovi_positivi,dimessi_guariti,deceduti,casi_da_sospetto_diagnostico,casi_da_screening,totale_casi,tamponi,casi_testati,note,ingressi_terapia_intensiva,note_test,note_casi,totale_positivi_test_molecolare,totale_positivi_test_antigenico_rapido,tamponi_test_molecolare,tamponi_test_antigenico_rapido
2020-02-24T18:00:00,ITA,101,26,127,94,221,0,221,1,7,,,229,4324,,,,,,,,,
2020-02-25T18:00:00,ITA,114,35,150,162,311,90,93,1,10,,,322,8623,,,,,,,,,
2020-02-26T18:00:00,ITA,128,36,164,221,385,74,78,3,12,,,400,9587,,,,,,,,,
2020-02-27T18:00:00,ITA,248,56,304,284,588,203,250,45,17,,,650,12014,,,,,,,,,
2020-02-28T18:00:00,ITA,345,64,409,412,821,233,238,46,21,,,888,15695,,,,,,,,,
2020-02-29T18:00:00,ITA,401,105,506,543,1049,228,240,50,29,,,1128,18661,,,,,,,,,
2020-03-01T18:00:00,ITA,639,140,779,798,1577,528,566,83,34,,,1694,21127,,,,,,,,,
2020-03-02T18:00:00,ITA,742,166,908,927,1835,258,342,149,52,,,2036,23345,,,,,,,,,
2020-03-03T18:00:00,ITA,1034,229,1263,1000,2263,428,466,160,79,,,2502,25856,,,,,,,,,
Remember to change Cote d'Ivoire to Cote d_Ivoire in order to avoid parsing CSV error:
$ sed -ie s/d\'/d_/g country_vaccinations.csv
--
| name | type | num_values | distinct_count | total_count | missing_count | int_count | min | max |
|---|---|---|---|---|---|---|---|---|
| country | nominal | 223 | 223 | 71815 | 0 | 71815 | 0.0 | 0.0 |
| iso_code | nominal | 223 | 223 | 71815 | 0 | 71815 | 0.0 | 0.0 |
| date | date | 0 | 415 | 71815 | 0 | 0 | 1.6067772E12 | 1.6425468E12 |
| total_vaccinations | numeric | 0 | 36847 | 71815 | 34296 | 37389 | 0.0 | 2.951846E9 |
| people_vaccinated | numeric | 0 | 34781 | 71815 | 36098 | 35717 | 0.0 | 1.263691E9 |
| people_fully_vaccinated | numeric | 0 | 31730 | 71815 | 38880 | 32935 | 1.0 | 1.220584E9 |
| daily_vaccinations_raw | numeric | 0 | 24889 | 71815 | 41195 | 30620 | 0.0 | 2.4741E7 |
| daily_vaccinations | numeric | 0 | 35706 | 71815 | 363 | 71452 | 0.0 | 2.2424286E7 |
| total_vaccinations_per_hundred | numeric | 0 | 15374 | 71815 | 34296 | 608 | 0.0 | 325.99 |
| people_vaccinated_per_hundred | numeric | 0 | 8651 | 71815 | 36098 | 589 | 0.0 | 122.49 |
| people_fully_vaccinated_per_hundred | numeric | 0 | 8179 | 71815 | 38880 | 874 | 0.0 | 119.62 |
| daily_vaccinations_per_million | numeric | 0 | 12213 | 71815 | 363 | 71452 | 0.0 | 117497.0 |
| vaccines | nominal | 78 | 78 | 71815 | 0 | 71815 | 0.0 | 0.0 |
| source_name | nominal | 83 | 83 | 71815 | 0 | 71815 | 0.0 | 0.0 |
| source_website | nominal | 130 | 130 | 71815 | 694 | 71121 | 0.0 | 0.0 |
head ../data/country_vaccinations.csv
country,iso_code,date,total_vaccinations,people_vaccinated,people_fully_vaccinated,daily_vaccinations_raw,daily_vaccinations,total_vaccinations_per_hundred,people_vaccinated_per_hundred,people_fully_vaccinated_per_hundred,daily_vaccinations_per_million,vaccines,source_name,source_website
Afghanistan,AFG,2021-02-22,0.0,0.0,,,,0.0,0.0,,,"Johnson&Johnson, Oxford/AstraZeneca, Pfizer/BioNTech, Sinopharm/Beijing",World Health Organization,https://covid19.who.int/
Afghanistan,AFG,2021-02-23,,,,,1367.0,,,,34.0,"Johnson&Johnson, Oxford/AstraZeneca, Pfizer/BioNTech, Sinopharm/Beijing",World Health Organization,https://covid19.who.int/
Afghanistan,AFG,2021-02-24,,,,,1367.0,,,,34.0,"Johnson&Johnson, Oxford/AstraZeneca, Pfizer/BioNTech, Sinopharm/Beijing",World Health Organization,https://covid19.who.int/
Afghanistan,AFG,2021-02-25,,,,,1367.0,,,,34.0,"Johnson&Johnson, Oxford/AstraZeneca, Pfizer/BioNTech, Sinopharm/Beijing",World Health Organization,https://covid19.who.int/
Afghanistan,AFG,2021-02-26,,,,,1367.0,,,,34.0,"Johnson&Johnson, Oxford/AstraZeneca, Pfizer/BioNTech, Sinopharm/Beijing",World Health Organization,https://covid19.who.int/
Afghanistan,AFG,2021-02-27,,,,,1367.0,,,,34.0,"Johnson&Johnson, Oxford/AstraZeneca, Pfizer/BioNTech, Sinopharm/Beijing",World Health Organization,https://covid19.who.int/
Afghanistan,AFG,2021-02-28,8200.0,8200.0,,,1367.0,0.02,0.02,,34.0,"Johnson&Johnson, Oxford/AstraZeneca, Pfizer/BioNTech, Sinopharm/Beijing",World Health Organization,https://covid19.who.int/
Afghanistan,AFG,2021-03-01,,,,,1580.0,,,,40.0,"Johnson&Johnson, Oxford/AstraZeneca, Pfizer/BioNTech, Sinopharm/Beijing",World Health Organization,https://covid19.who.int/
Afghanistan,AFG,2021-03-02,,,,,1794.0,,,,45.0,"Johnson&Johnson, Oxford/AstraZeneca, Pfizer/BioNTech, Sinopharm/Beijing",World Health Organization,https://covid19.who.int/
| name | type | num_values | distinct_count | total_count | missing_count | int_count | min | max |
|---|---|---|---|---|---|---|---|---|
| location | nominal | 40 | 40 | 25783 | 0 | 25783 | 0.0 | 0.0 |
| date | date | 0 | 403 | 25783 | 0 | 0 | 1.6070364E12 | 1.6425468E12 |
| vaccine | nominal | 8 | 8 | 25783 | 0 | 25783 | 0.0 | 0.0 |
| total_vaccinations | numeric | 0 | 22830 | 25783 | 0 | 25783 | 0.0 | 5.47608975E8 |
head ../data/country_vaccinations_by_manufacturer.csv
location,date,vaccine,total_vaccinations
Austria,2021-01-08,Johnson&Johnson,0
Austria,2021-01-08,Moderna,0
Austria,2021-01-08,Oxford/AstraZeneca,0
Austria,2021-01-08,Pfizer/BioNTech,31513
Austria,2021-01-15,Johnson&Johnson,0
Austria,2021-01-15,Moderna,95
Austria,2021-01-15,Oxford/AstraZeneca,0
Austria,2021-01-15,Pfizer/BioNTech,116840
Austria,2021-01-22,Johnson&Johnson,0
- Type - Ordered Temporal Data
- Missing data: true
- Duplicate data: false
Preparation
Dataset: The country_vaccinations_by_manufacturer dataset have all of needed data to match the goal.
Strategy: Transform data in pivot table, order desc and select top 10 country.
| Location | Pfizer/BioNTech | Moderna | Oxford/AstraZeneca | Sinovac | Johnson&Johnson | Sinopharm/Beijing | Sputnik V | CanSino |
|---|---|---|---|---|---|---|---|---|
| European Union | 547 | 125 | 67 | 0 | 18 | 2 | 1 | 0 |
| United States | 310 | 201 | 0 | 0 | 18 | 0 | 0 | 0 |
| Germany | 117 | 26 | 12 | 0 | 3 | 0 | 0 | 0 |
| France | 102 | 21 | 7 | 0 | 1 | 0 | 0 | 0 |
| Italy | 80 | 27 | 12 | 0 | 1 | 0 | 0 | 0 |
| South Korea | 65 | 21 | 22 | 0 | 1 | 0 | 0 | 0 |
| Japan | 86 | 5 | 0 | 0 | 0 | 0 | 0 | 0 |
| Spain | 55 | 18 | 9 | 0 | 1 | 0 | 0 | 0 |
| Peru | 30 | 0 | 4 | 0 | 0 | 19 | 0 | 0 |
| Poland | 35 | 3 | 5 | 0 | 2 | 0 | 0 | 0 |
Goal: Try to predict the spread of COVID-19 ahead of time to take preventive measures (Italy subset)
Preparation
Dataset: The dpc-covid19-ita-andamento-nazionale dataset have all of needed data to match the goal.
Strategy: Repair missing values Remove unused attribute Add new attribute: tasso_positivita = 100 * nuovi_positivi / Tamponi delta in percent Predict trend for 100 gg ahead with different algorithms
measure elapsed time (wall clock) in ms
| algorithm_name | elapsed (ms) |
|---|---|
| GaussianProcesses | 1153 |
| MultilayerPerceptron | 1487 |
| LinearRegression | 134 |
| SMOreg | 437 |
- the graph was produced by google sheet chart
- SMO regression may be not appropriate for this use case?
Hello Weka!
Working Directory: /Users/nick/eclipse-workspace/weka-covid19
**********************************
Data set name: dpc-covid19-ita-andamento-nazionale
Data set size: 695
See res/dpc-covid19-ita-andamento-nazionale.attribute.md
**********************************
Data set name: country_vaccinations
Data set size: 71815
See res/country_vaccinations.attribute.md
**********************************
Data set name: country_vaccinations_by_manufacturer
Data set size: 25783
See res/country_vaccinations_by_manufacturer.attribute.md
**********************************
pivot
See res/country_vaccinations_by_manufacturer.top.md
See res/country_vaccinations_by_manufacturer.top.csv
**********************************
join
We can do join!!!
See res/join.csv
Data set load: res/join.csv
**********************************
Data set name: join
Data set size: 386
See res/join.attribute.md
**********************************
predict
See ./res/join.arff
See ./res/forecast.csv
See ./res/cost.md
Bye-bye Weka!