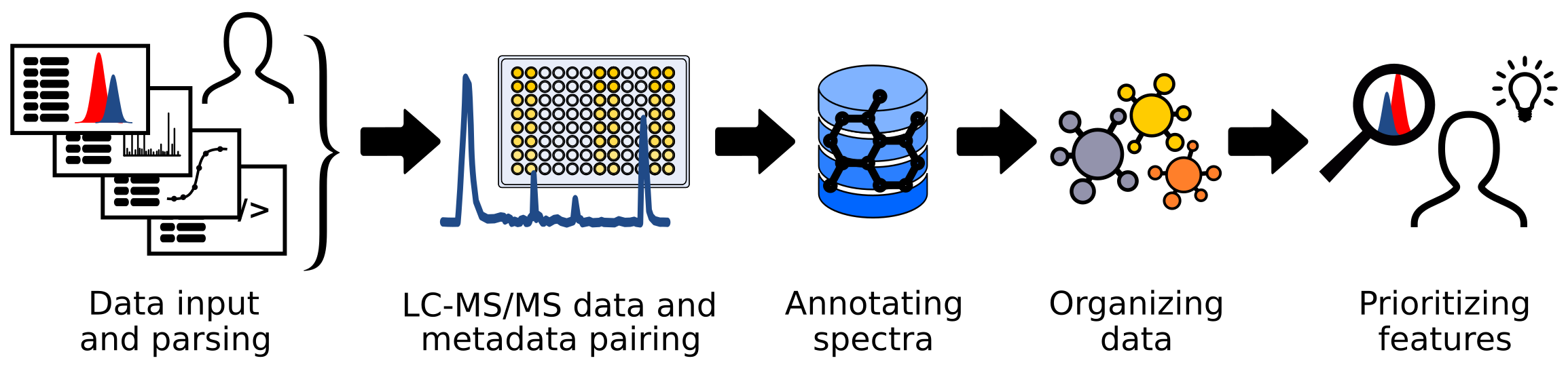
FERMO is a dashboard app for the processing and visualization of liquid chromatography - (tandem) mass spectrometry data, and its pairing to quantitative biological data and other metadata. In particular, FERMO is aimed toward the prioritization of compounds putatively responsible for a biological observation, for example investigating molecules that may be responsible for antibiotic activity. Developed with natural products research in mind, FERMO can be also used in other areas of the life sciences, such as metabolomics or environmental sciences, to investigate metabolites, drugs, pollutants, or agrochemicals.
More information on the software can be found in the preprint on biorXiv: Mitja M. Zdouc et al. FERMO: a Dashboard for Streamlined Rationalized Prioritization of Molecular Features from Mass Spectrometry Data. bioRxiv (2022).
References, tutorials, and guides can be found in the FERMO Github Wiki.
Of note, FERMO is currently in beta development. If you notice a bug or another issue, please contact us by opening an issue on FERMO's Github page.
Although FERMO uses a browser (e.g. Firefox, Google Chrome, Microsoft Edge, ...) to render the graphical user interface of the application, it runs fully local. No internet connection is required except during the installation, and no data is shared across the internet.
FERMO requires untargeted high resolution data-dependend acquisition liquid chromatography tandem mass spectrometry (HR-DDA-LC-MS/MS) data to work properly. Currently, annotation is restricted to positive ion mode data.
FERMO needs the following prerequisites:
- Write permissions on the computer
- Python version 3.8
- Python package manager (recommended, tested with Miniconda3/Anaconda3)
- Python dependencies (see below for a list of required packages)
FERMO performs the analysis with following input files:
- Mandatory files for analysis:
- a peak table in MZmine 3 'quant_full.csv' format (tested with MZmine3 versions 3.0.0 - 3.3.0)
- the accompanying .mgf-file
- Optional files for analysis:
- a .csv file containing group metadata information
- a .csv file containing quantitative biological data information
- a .mgf file containing a targeted spectral library
Example files can be found in the example_data folder. Instructions for the generation of the analysis files, as well as the required format, can be found in the user guides on input data formats in the FERMO GitHub Wiki.
FERMO must be downloaded and installed before use. This can be done by simply downloading this repository as ZIP-compressed folder (via the green "Code" button on the top right side of this page -> "Download ZIP"), or by cloning it. For the installation, see the step-by-step instructions below (Windows, Linux).
-
Download FERMO to an easily accessible location with write permissions, such as the Downloads folder or the Desktop. Unpack the package, for example by using 7-Zip . Alternatively, clone this repository.
-
Download the latest Miniconda3 version from the Conda website and install it with default settings. Details on the installation procedure can be found in this Guide. Anaconda can also be used, but its installation requires substantially more disk space. If Miniconda or Anaconda were already installed before, please update to the latest version, to prevent errors during the installation. Of note, other package managers may be used, but have not been tested.
-
Once Miniconda3/Anaconda3 was installed/updated, there are two options to run FERMO. The easiest option is by double-clicking the startup script
FERMO_START_WINDOWS.batin the FERMO directory. This script will start FERMO, create a package management environment, download and install all dependencies, and open a browser window where FERMO will already be running. -
If the script was not able to install and/or start FERMO, it might be because Miniconda3/Anaconda3 was installed on another drive than
C:. In this case, start FERMO manually as indicated in step 5. Other commonly observed errors and their solutions are described int the FAQs. -
Manual installation and start of FERMO: Open the Anaconda Prompt from the Windows Start menu. The following steps must be performed in the Anaconda prompt command line window.
-
Navigate to the previously downloaded and unpacked FERMO directory, changing directories with the command
cdand showing the contents of the current directory with the commanddir. This must be done every time FERMO is started. -
In the FERMO directory, create a Conda virtual environment by entering the command
conda create --name FERMO python=3.8. This must be done only the first time FERMO is run. -
Activate the newly created virtual environment by entering the command
conda activate FERMO. The prefix of the command prompt should switch to(FERMO), to indicate the change in environment. This must be done every time FERMO is started. -
In the FERMO directory, install the dependencies by running the command
pip install -e .(take care to also copy the trailing dot). -
Start FERMO with the command
python ./src/fermo/app.py. After a few seconds, a browser window will automatically open, running FERMO on the local IP address127.0.0.1:8050. -
After use, close the command line window in which FERMO runs, or terminate execution of the program by hitting
ctrl+c.
Installation on macOS is currently not possible. We are working on fixing the issue.
-
Download FERMO to an easily accessible location with write permissions, such as the Downloads folder or the Desktop. Unpack the package. Alternatively, clone this repository.
-
Download the latest Miniconda3 version from the Conda website and install it with default settings. Details on the installation procedure can be found in this Guide. Anaconda can also be used, but its installation requires substantially more disk space. If Miniconda or Anaconda were already installed before, please update to the latest version, to prevent errors during the installation. Of note, other package managers may be used, but have not been tested with FERMO.
-
Once Miniconda3/Anaconda3 was installed/updated, close and re-open the terminal window to make changes take effect. The terminal prompt should now have the prefix
(base). -
FERMO can be started in two ways. The easiest option is double-clicking the startup script
FERMO_START_LINUX.sh, which can be found in the FERMO directory. This script will start FERMO, create a conda environment, activate it, install all dependencies, and open a browser window where FERMO will already be running. However, on some Linux distributions, shell-scripts cannot be opened in the file explorer for security reasons. In this case, open a new terminal window, navigate to the FERMO directory, and execute the commandschmod +x ./FERMO_START_LINUX.shand./FERMO_START_LINUX.sh. If that does not work, FERMO can also be started manually, as indicated in the next step. Other commonly observed errors and their solutions are described int the FAQs. -
Manual installation and start of FERMO: Open a new terminal window. Navigate to the previously downloaded and unpacked FERMO directory, using the
cd("change directory") andls("list directory") commands. This must be done every time FERMO is started. -
In the FERMO directory, create a virtual environment by entering the command
conda create --name FERMO python=3.8. This must be done only at the first start of FERMO. -
Activate the newly created virtual environment by entering the command
conda activate FERMO. The prefix of the command prompt should switch to(FERMO), to indicate the change in environment. This must be done whenever FERMO is started. -
In the FERMO directory, install the dependencies by running the command
pip install -e .(take care to also copy the trailing dot). -
To start FERMO, enter the command
python ./src/fermo/app.py. After a few seconds, a browser window will automatically open, running FERMO on the local IP address127.0.0.1:8050. -
After use, close the command line window in which FERMO runs, or terminate execution of the program by hitting
ctrl+c.
- Sample: a contiguous instrument run containing MS1 (surveying) scans and or MS2 (collision cell) scans.
- Molecular feature: an LC-MS extracted ion chromatogram (EIC) peak with specific attributes (e.g. mass-to-charge-ratio m/z, retention time RT, ...).
- Peak table: a table summarizing detected molecular features across samples.
This guide describes the essential steps to process and analyze data with FERMO, using files from the example data described in the article Mitja M. Zdouc et al. FERMO: a Dashboard for Streamlined Rationalized Prioritization of Molecular Features from Mass Spectrometry Data. bioRxiv (2022). A more thorough guide, including information on scores and data processing, can be found in the FERMO Github Wiki.
-
Start FERMO, following the instructions in the installation guide above.
-
On the landing page, select the Processing mode.
-
On the Processing mode page, load the files by clicking on the respective buttons:
- Load peak table -> "case_study_peak_table_quant_full.csv"
- Load the MS/MS file -> "case_study_MSMS.mgf"
- Specify format of quantitative biological data -> "Percentage-like"
- Load quantitative biological data -> "case_study_bioactivity.csv"
- Load group metadata table -> "case_study_group_metadata.csv"
- Load spectral library -> "case_study_spectral_library.mgf"
-
(Optional) on the Processing mode page, set the "MS2Query" parameter on the right-hand side to "ON".
-
Start FERMO by clicking the 'Start FERMO' button at the bottom of the page. Processing will take a few minutes. If MS2Query annotation was selected, library files are automatically downloaded from Zenodo . Due to their size, this can take a while. Once processing is finished, the dashboard will load automatically.
-
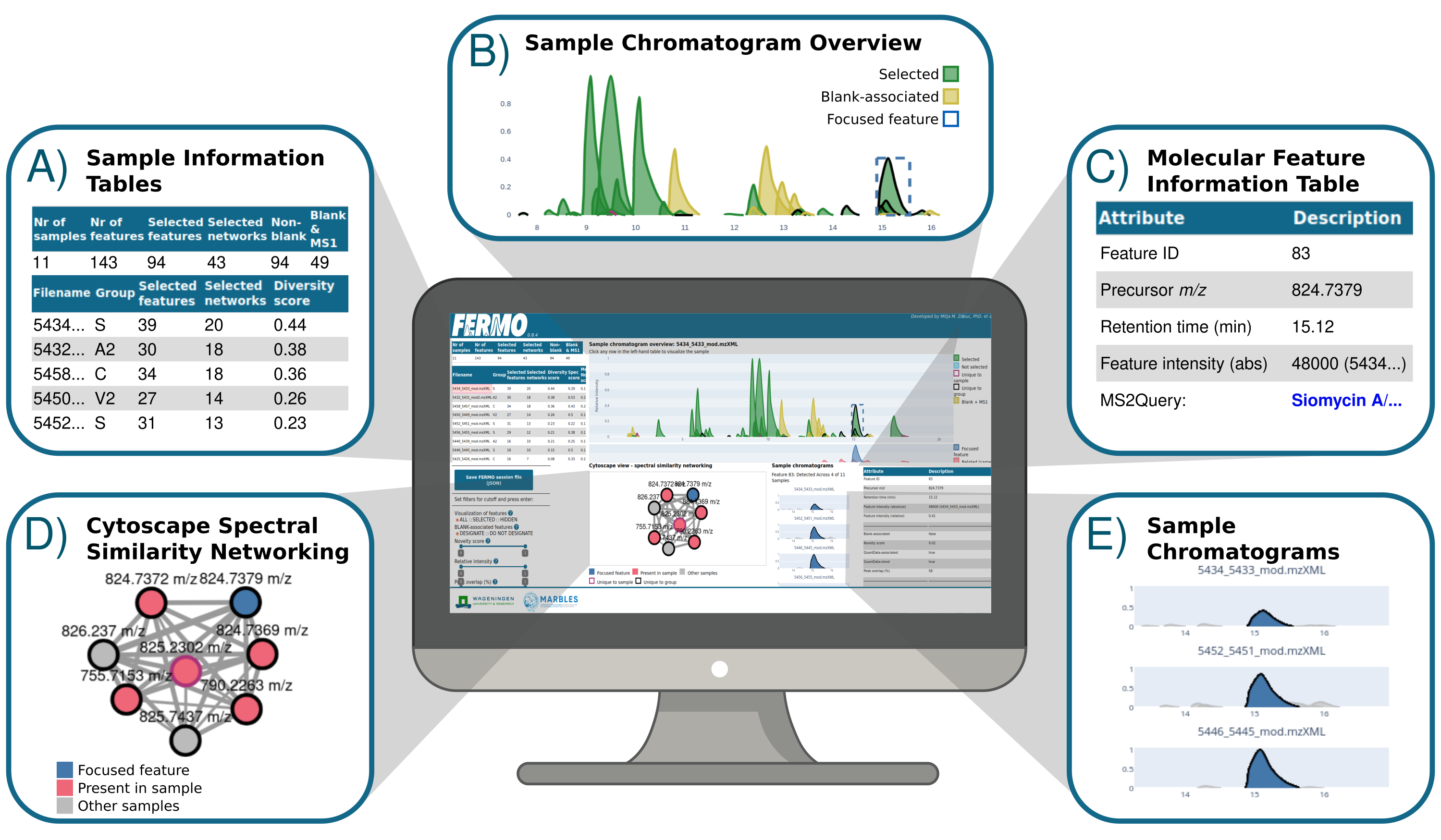
The dashboard is organized into six fields:
- Sample Information Tables (A)
- Sample Chromatogram Overview (B)
- Molecular Feature Information Table (C)
- Cytoscape Spectral Similarity Networking (D)
- Sample Chromatograms (E)
- Filter and Export Panel
-
To display a specific sample, click on one of the rows in the Sample Information Tables (A). This triggers the update of the Sample Chromatogram Overview (B), which displays the molecular features detected in the sample. Hovering over a molecular feature presents general information. A click on a molecular feature focuses it, which triggers an update of the Molecular Feature Information Table (C), the Cytoscape Spectral Similarity Networking (D), and the Sample Chromatograms (E).
-
In the Molecular Feature Information Table (C), all information about the focused molecular feature is displayed. This includes its general attributes, its calculated scores, its annotations, and information about its similarity network.
-
In the Sample Chromatograms (E) view, the distribution of the focused molecular feature across samples is displayed, which can be used to determine the sample most suitable for further exploration (e.g. chromatographic isolation).
-
The Cytoscape Spectral Similarity Networking (D) shows the relationship between molecular features, based on similarity of their MS/MS spectra. Clicking nodes or edges triggers the node information table or edge information table, respectively, which are below the network view.
-
In the Filter and Export Panel, different filters can be set that change the current selection of molecular features. For example, to find all molecular features annotated as siomycin, enter "siomycin" into the "Annotation search" field and press enter. Filters can be also combined with each other, to generate more complex selection conditions.
A list of dependencies can be found in requirements.txt.
FERMO is licensed under the MIT License - see the LICENSE.md file for details.
For details on our code of conduct and the process for submitting pull requests to us, please read CONTRIBUTING.md.
We use Semantic Versioning for versioning.
- Mitja M. Zdouc - Lead developer -
- Hannah E. Augustijn - Design -
See also the list of contributors who participated in this project.
- Bayona Maldonado, Lina M. - beta testing
- Dell, Maria - beta testing
- van der Hooft, Justin J. J. - project supervision
- Iorio, Marianna - beta testing
- de Jonge, Niek - code review, beta testing
- Khatib, Soliman - beta testing
- Lund, George - beta testing
- Maffioli, Sonia - beta testing
- Medema, Marnix H. - project supervision
- Padva, Leo - beta testing
- Simone, Matteo - beta testing
- Soldatou, Sylvia - beta testing