This repository includes code to train a simple RF classifier to predict the origin of silvereyes (Zosterops lateralis) based on their morphology. The
.
├── data # Data used in the project
│ └── raw # Original, unmodified data
│ └── morph-df.csv # CSV file with raw data
├── environment.yml # Conda environment file
├── figures # Output figures from the analysis
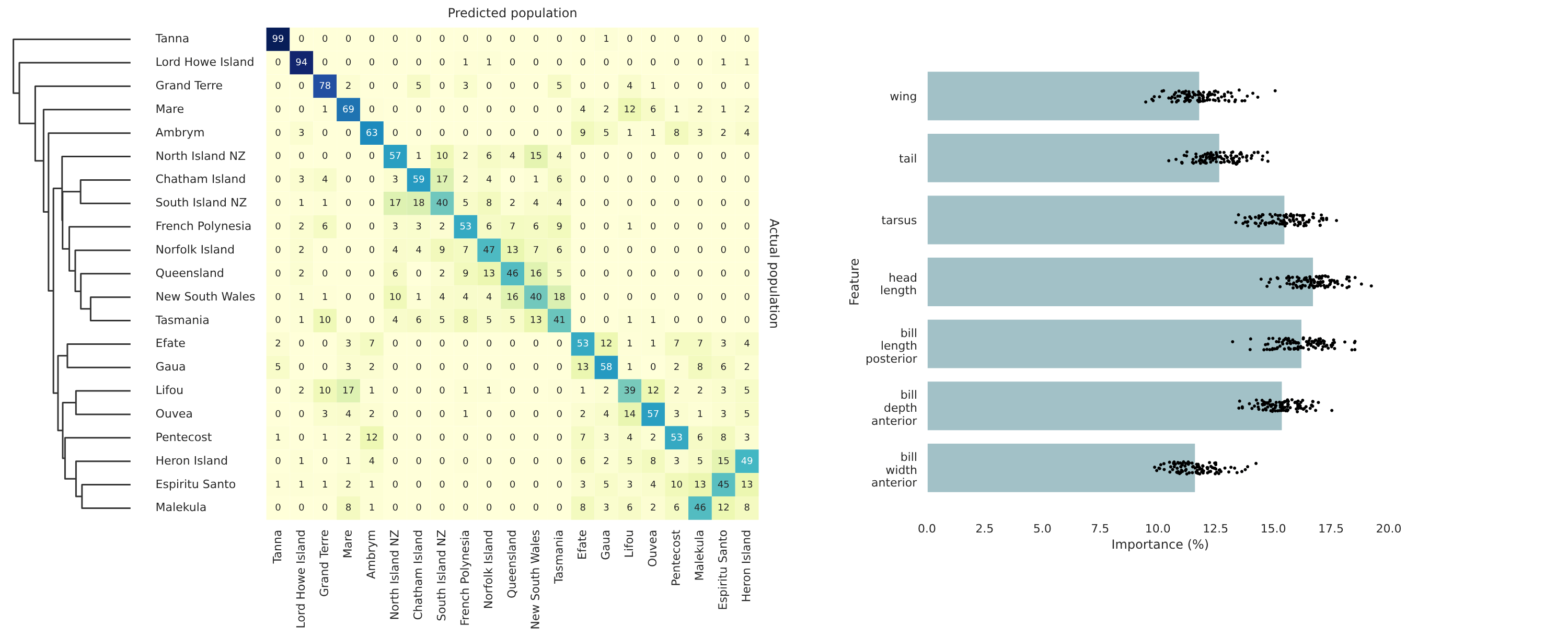
│ ├── confusion-matrix-clustermap.pdf # Clustermap of the confusion matrix
│ ├── confusion-matrix.pdf # Confusion matrix
│ ├── feature-importances.pdf # Plot of feature importances
│ └── permutation-test-score.pdf # Permutation test score plot
├── README.md # The file you're reading now
├── scripts # Scripts to reproduce the analysis
│ └── train-rf.py # Script to train the random forest model
└── src # Source code for the project
This project uses a specific Conda environment. To reproduce this environment on your machine, follow these steps:
git clone https://github.com/nilomr/silvereye-morphology.git-
Navigate to the repository's directory.
-
Run the following command to create the Conda environment from the
environment.ymlfile:
conda env create -f environment.yml- Activate the environment:
conda activate silvereye-morphologyThe train-rf.py script is used to train the random forest model. Here's how to run it:
Make sure your Conda environment is activated, then navigate to the scripts
directory, and run the following command:
python train-rf.py
The script will output a heatmap plot as a PDF file.
If you encounter any bugs or issues, please let me know by creating an issue in this repository.
© Nilo M. Recalde, 2023