Code repository for 2023 MICCAI workshop FAIMI paper: Are Sex-based Physiological Differences the Cause of Gender Bias for Chest X-ray Diagnosis?
The code repo should be functioning well under python==3.9.11.
Create a new conda enviroment:
conda create -n detecting_causes python=3.9.11
Install the packages in requirements.txt:
conda install -f requirements.txt
Or: (prob work better)
conda env create -f env.yml
Both datasets are available online.
For a easy way to download it:
- NIH: link. Download it and unzip it based on its instructions.
- CheXpert: link.
- A smaller version on Kaggle: link: resized image. Which does not matter that much for this work, as we will resize it to 224 x 224 when preprocessing. Need to mention here that resizing the image will definitely influence the prediction ability; however, in this work, we care more about the performance gap between groups rather than getting better performance.
- Notice that we only use the frontal images (include PA and AP view position) in this work.
Mata data has already been processed, please refer to chexpert.sample.allrace.csv (CheXpert) and
Data_Entry_2017_v2020_clean_cplit.csv (NIH).
run the following command to pre-process the images (resize).
python3 ./preprocess/preprocimgs.py -p {your dataset path}
Command example for training NIH dataset on label Pneumothorax with resampling strategy with random state from 0 to 10, 0, 50 and 100 female percentage in training:
First, go to the predction folder:
cd ./prediction/
python3 ./disease_prediction.py -s NIH -d Pneumothorax -f 0 50 100 -n 1 -r 0-10 -p {your dataset folder path}
Train on CheXpert:
python3 ./disease_prediction.py -s chexpert -d Pneumothorax -f 0 50 100 -n 1 -r 0-10 -p {your dataset folder path}
Train on different disease labels sequently:
python3 ./disease_prediction.py -s NIH -d Pneumothorax Pneumonia Cadiomegaly -f 0 50 100 -n 1 -r 0-10 -p {your dataset folder path}
Details about the other hyper-parameters could be found in the same py file.
Sampling actually takes quite a time:
#TODO: prepare the splits for different random states as csv in ./datafiles/
For plotting all the disease after re-store the results into csv file:
python3 ./analysis/plotting.py -p {your run result dir path}
If you train the model with changed hyper-parameters rather then the required 5 (-s -d -f -n -r), you need to add another hyperparameter mannually as -f.
For example:
python3 ./analysis/plotting.py -p {your run result dir path} -f -image_size1024
when set --image_size 1024 during training.
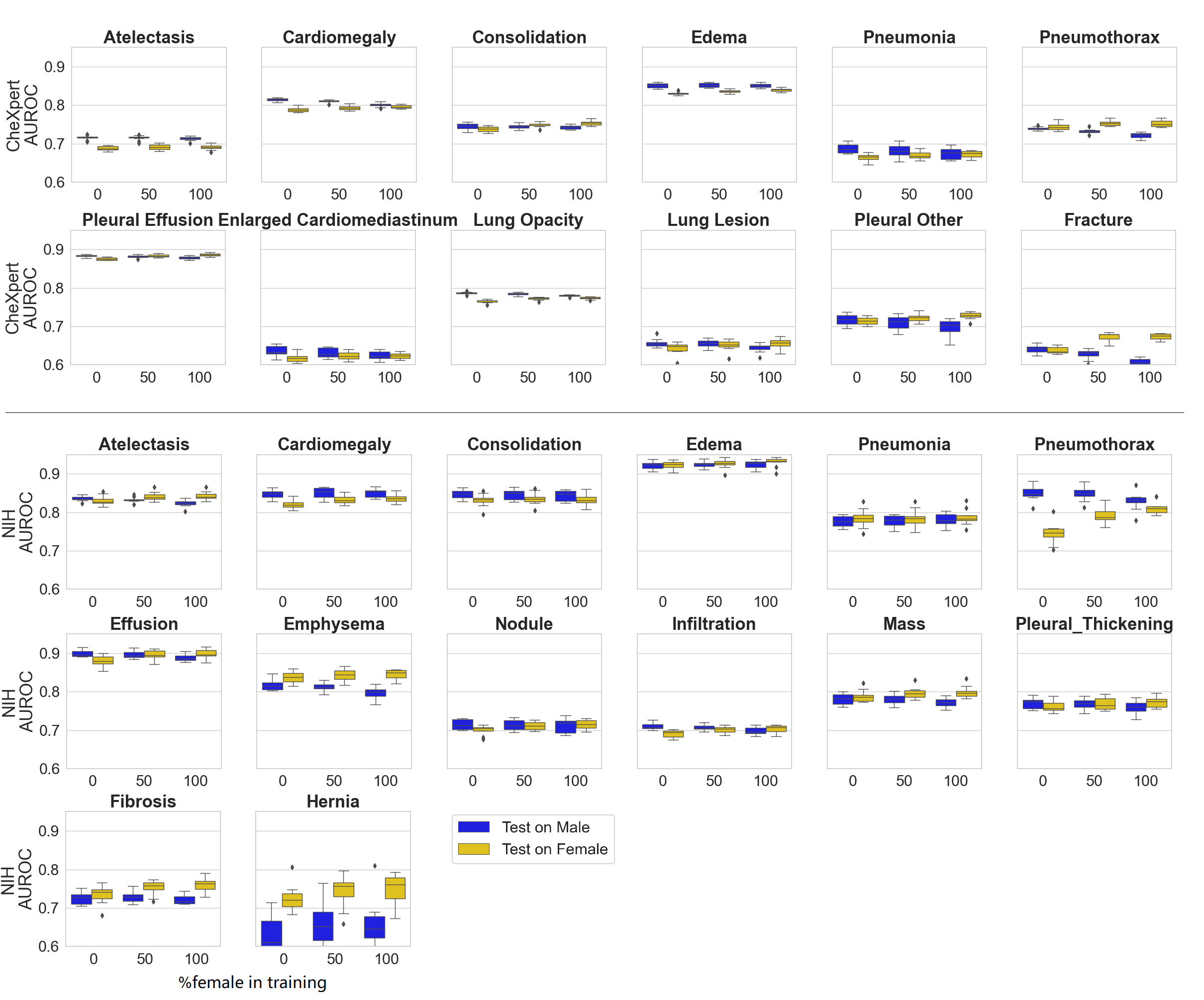
(Well, you need to edit the image a bit to make it like this;))
#TODO When specify dataset as ['NIH','chexpert'], the plotting has index errors. Fix it!!
See above Plotting the results.
To run the experiments with cropped images:
python3 ./prediction/disease_prediction.py -s NIH -d Pneumothorax -f 0 50 100 -n 1 -r 0-10 --crop 0.6 -p {your dataset folder path}
Notice that this part of experiment does not apply to CheXpert.
- Without prioritizing the diseased one: #TODO
- Without sampling or sample more than one scans per patient: change the
-nhyper-parameter:
python3 ./prediction/disease_prediction.py -s NIH -d Pneumothorax -f 0 50 100 -n None -r 0-10 -p {your dataset folder path} - Change the prevalence setting:
--prevalence_setting totalor--prevalence_setting equal
- Save model parameters:
python3 ./prediction/disease_prediction.py -s NIH -d Pneumothorax -f 0 50 100 -n None -r 0-10 -p {your dataset folder path} --save_model True - Run the cross dataset inference:
python3 ./analysis/cross_ds_inference.py -d Penumothorax --run_dir {your run dir} --data_dir {your dataset dir}