Code for our paper "A Bio-Inspired Multi-Exposure Fusion Framework for Low-light Image Enhancement"
- The code for the comparison method is also provided, see lowlight
- Downloads: google Drive (Just unzip data to current folder)
- Datasets
VV, LIME, NPE, NPE-ex1, NPE-ex2, NPE-ex3, DICM, MEF - Since some methods are quite time-consuming, we also provide their results (e.g.
results__dong@VV.zip) - Since some metrics are quite time-consuming, we also provide their results (
TestReport.zip)
- Datasets
- All the experiments can be reproduced easily by running
experiments.m
From left to right: input images, results of MSRCR, Dong, NPE, LIME, MF, SRIE, and BIMEF(ours).
- VV (**Busting image enhancement and tone-mapping algorithms: **A collection of the most challenging cases)
- LIME-data
- NPE-data, NPE-ex1, NPE-ex2, NPE-ex3
- DICM —— 69 captured images from commercial digital cameras: Download (15.3 MB)
- MEF dataset
- Original code is tested on Matlab 2016b 64bit, Windows 10.
- matlabPyrTools is required to run VIF metric (
vif.m).
Run startup.m to add required path, then you are able to try the following demo.
I = imread('yellowlily.jpg');
J = BIMEF(I);
subplot 121; imshow(I); title('Original Image');
subplot 122; imshow(J); title('Enhanced Result');Replace BIMEF with other methods you want to test.
.
├── data # put your datasets here
│ ├── MEF # dataset name (VV, LIME, NPE...)
│ ├── out
│ │ ├── loe100x100 # LOE visualization results
│ │ ├── TestReport.csv # results of metrics
│ │ ├── TestReport__xxxx.csv # backups of TestReport
│ │ └── xxx__method.PNG # output images
│ └── xxx.jpg # input images
│
├── lowlight # lowlight image enhancement methods
├── quality # image quality metrics (blind or full-reference)
├── util # provide commonly used utility functions
│
├── demo.m # simple demo of lowlight enhancement
├── experiments.m # reproduce our experiments
└── startup.m # for installation
Run experiments.
% specify datasets
dataset = {'VV' 'LIME' 'NPE' 'NPE-ex1' 'NPE-ex2' 'NPE-ex3' 'MEF' 'DICM'};
dataset = strcat('data', filesep, dataset, filesep, '*.*');
% specify methods and metrics
method = {@multiscaleRetinex @dong @npe @lime @mf @srie @BIMEF};
metric = {@loe100x100 @vif};
for d = dataset, data = d{1};
data,
Test = TestImage(data);
Test.Method = method;
Test.Metric = metric;
% run test and display results
Test,
% save test to a .csv file
save(Test);
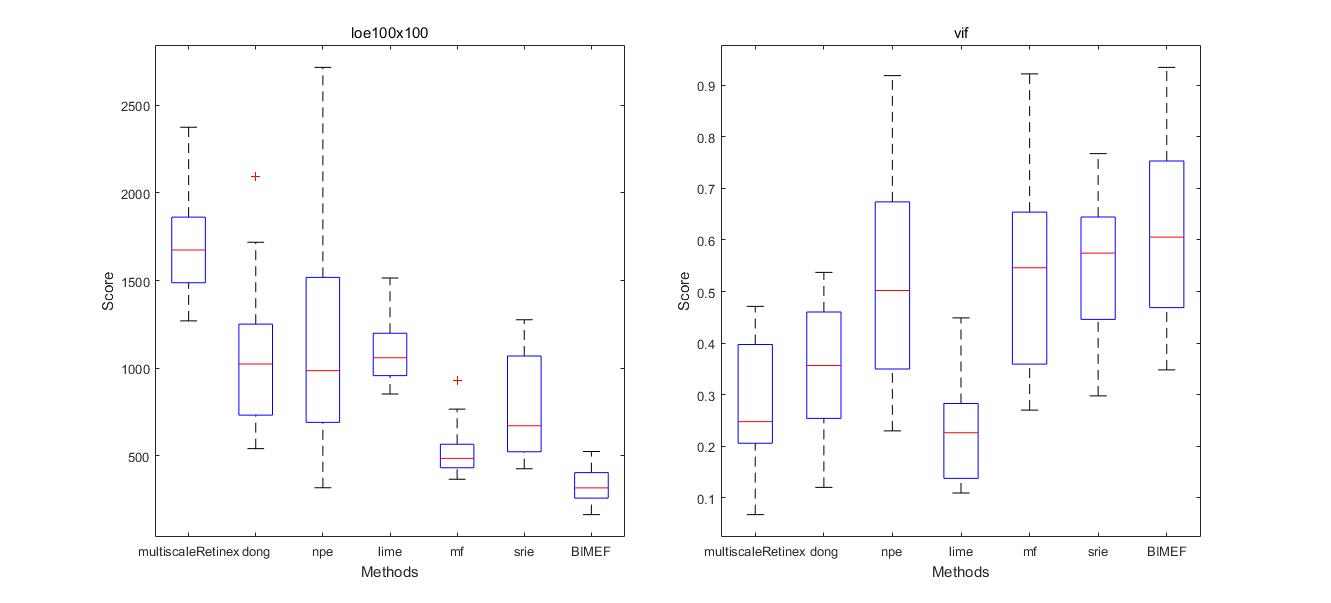
endShow test reports.
% table
TestReport('TestReport__VV.csv'),
% boxplot
TestReport('TestReport__MEF.csv').boxplot;Our method (BIMEF) has the lowest LOE and the highest VIF.