Run sh requirements.sh in a virtual environment in order to download the required libraries.
-
Open source biosignal acquisition hardware for research grade biosignal acquisition
-
Shirt using conductive textile electrodes [1$ for 1 electrode]
Wearable ECG electrodes --> OpenBCI Ganglion ---(Bluetooth)---> Raspberry Pi 4
- Ubuntu 16.04
- Nvidia 1080Ti - (Required for training the model)
- Download ECG MITDB monitoring data from https://storage.googleapis.com/mitdb-1.0.0.physionet.org/mit-bih-arrhythmia-database-1.0.0.zip and unzip it.
- To train:
python train.py --preprocess_data --data_path "PATH TO DATA"
- Download ECG from the BIDMC database which is derived from MIMIC-II from https://physionet.org/static/published-projects/bidmc/bidmc-ppg-and-respiration-dataset-1.0.0.zip and unzip it.
- To train:
python train.py --preprocess_data --data_path "PATH TO DATA"
- Download ECG from the preterm infant database from https://physionet.org/static/published-projects/picsdb/preterm-infant-cardio-respiratory-signals-database-1.0.0.zip and unzip it.
- Download the Heart Rate computation model from here: https://drive.google.com/open?id=1yI7G4nofjuzFWkD1CfsOtLZxaukTu0di
- Download the Breathing Rate computation model from here: https://drive.google.com/open?id=1ycV74LfGmgcGmLlrPn2VileeFNsGrRZT
- To run inference and view GUI type:
python run_model.py --path_dir "PATH TO DATA" --saved_hr_model_path "PATH TO HR MODEL" --saved_br_model_path "PATH TO BR MODEL" --patient_no 3 --viewer 1
Edge inference of ECG R-peak detection and Respiration extraction using Raspberry Pi 4 using ECG (OpenBCI Ganglion).
Wearable ECG electrodes --> OpenBCI Ganglion ---(Bluetooth)---> Raspberry Pi 4
OpenBCI client ----(LSL)---> Python -> PyTorch inference --> Breathing Rate, Heart Rate
- Install PyTorch dependicies
sudo apt install libopenblas-dev libblas-dev m4 cmake cython python3-yaml libatlas-base-dev - Increase swap file memory to 1600, Edit variable
CONF_SWAPSIZEin/etc/dphys-swapfile - Reset environmental variables like ONNX_ML Instructions
- Download PyTorch package compiled for Armv7 (torch-1.1.0-cp37-cp37m-linux_armv7l.whl)
- Install using the command
sudo pip3 install torch-1.1.0-cp37-cp37m-linux_armv7l.whlin the same directory
Refer here for troubleshooting
- Clone OpenBCI_Python repo
git clone htps://github.com/OpenBCI/OpenBCI_Python.git - Install the following requisites python packages using
pip3 installpylsl, python-osc, six, socketIO-client, websocket-client, Yapsy, xmldict, bluepy - Open folder OpenBCI_Python and run
sudo python3 user.py --board ganglion -a streamer_lslto open a lab streaming layer stream of sensor data from the ganglion
- Run the lsl streamer script to get data to the inference script
sudo python3 user.py --board ganglion -a streamer_lsl - Run the visualization and edge inference code on the pi using
python3 lsl_openbci.py
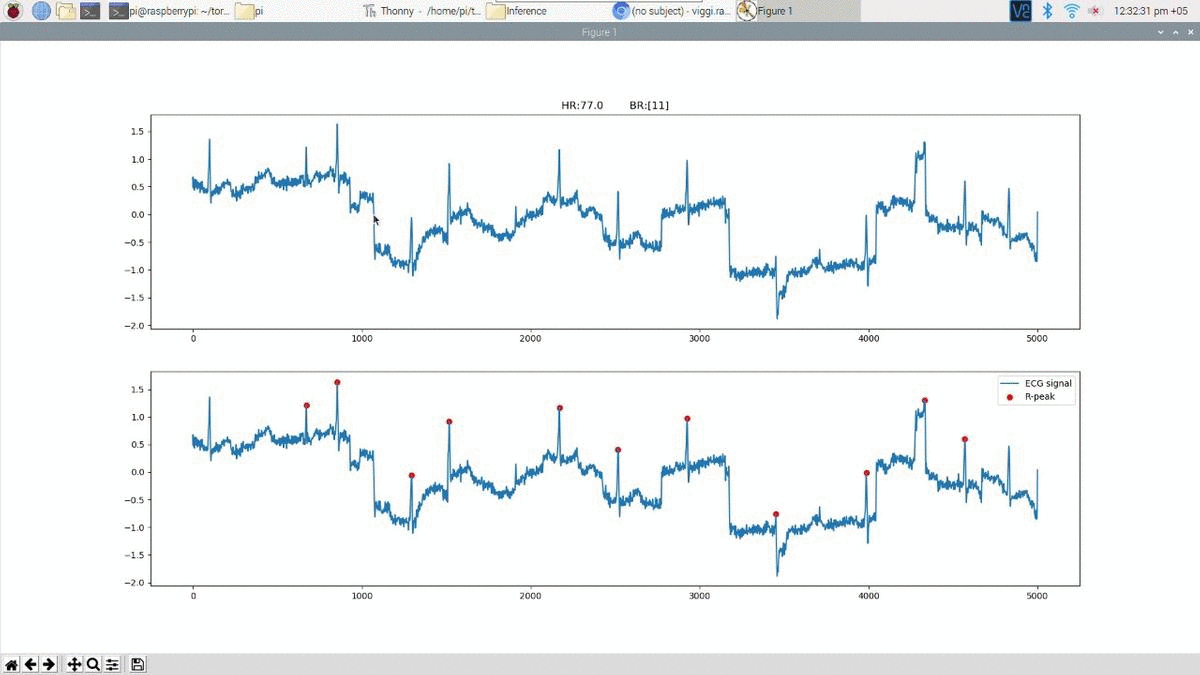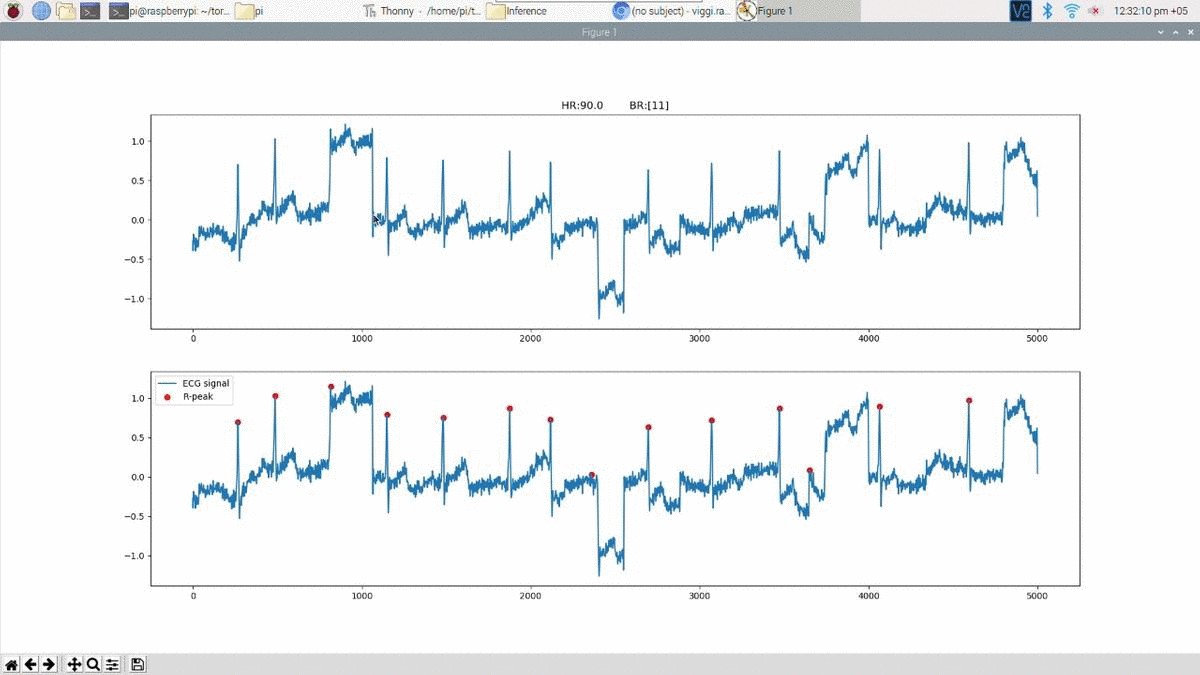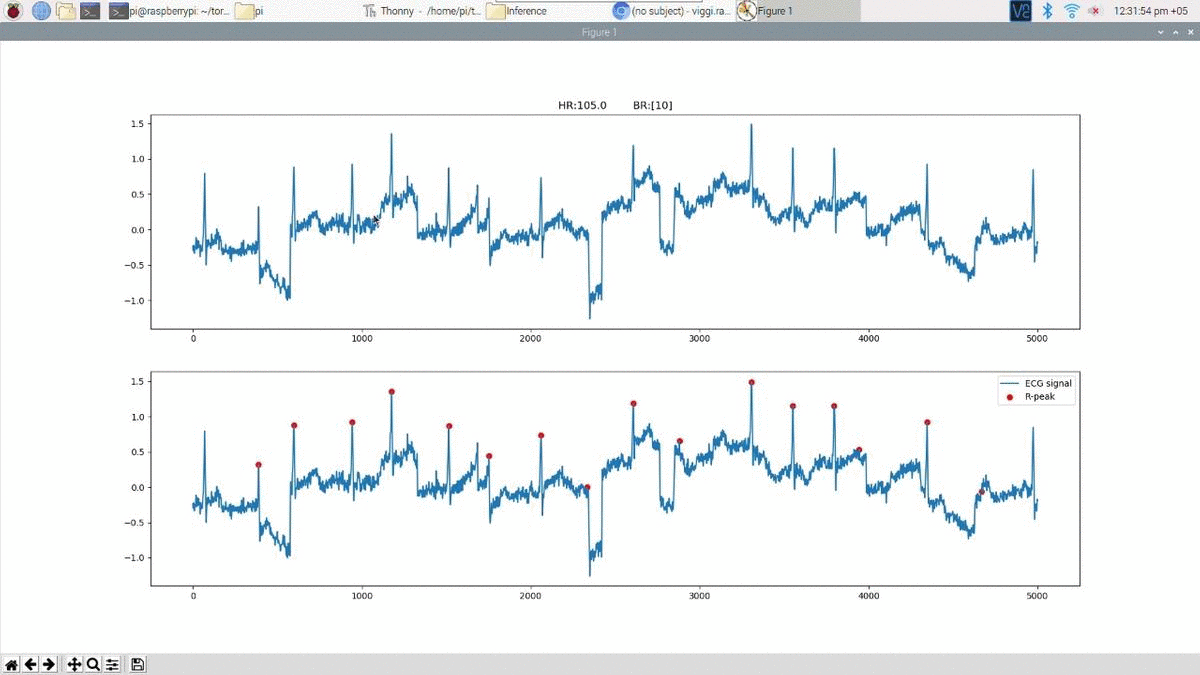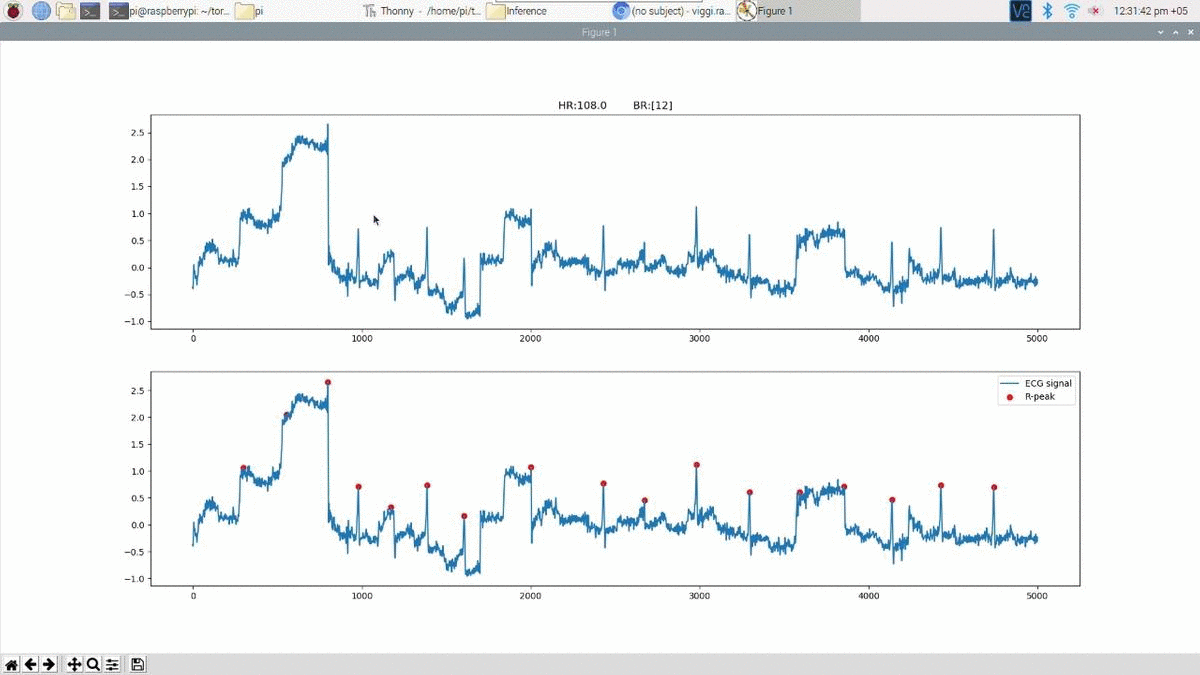 Above is an example prediction for noisy real time ECG data obtained using the edge inference model. The beat predictions are represented as blue markers on the ECG.
Above is an example prediction for noisy real time ECG data obtained using the edge inference model. The beat predictions are represented as blue markers on the ECG.


