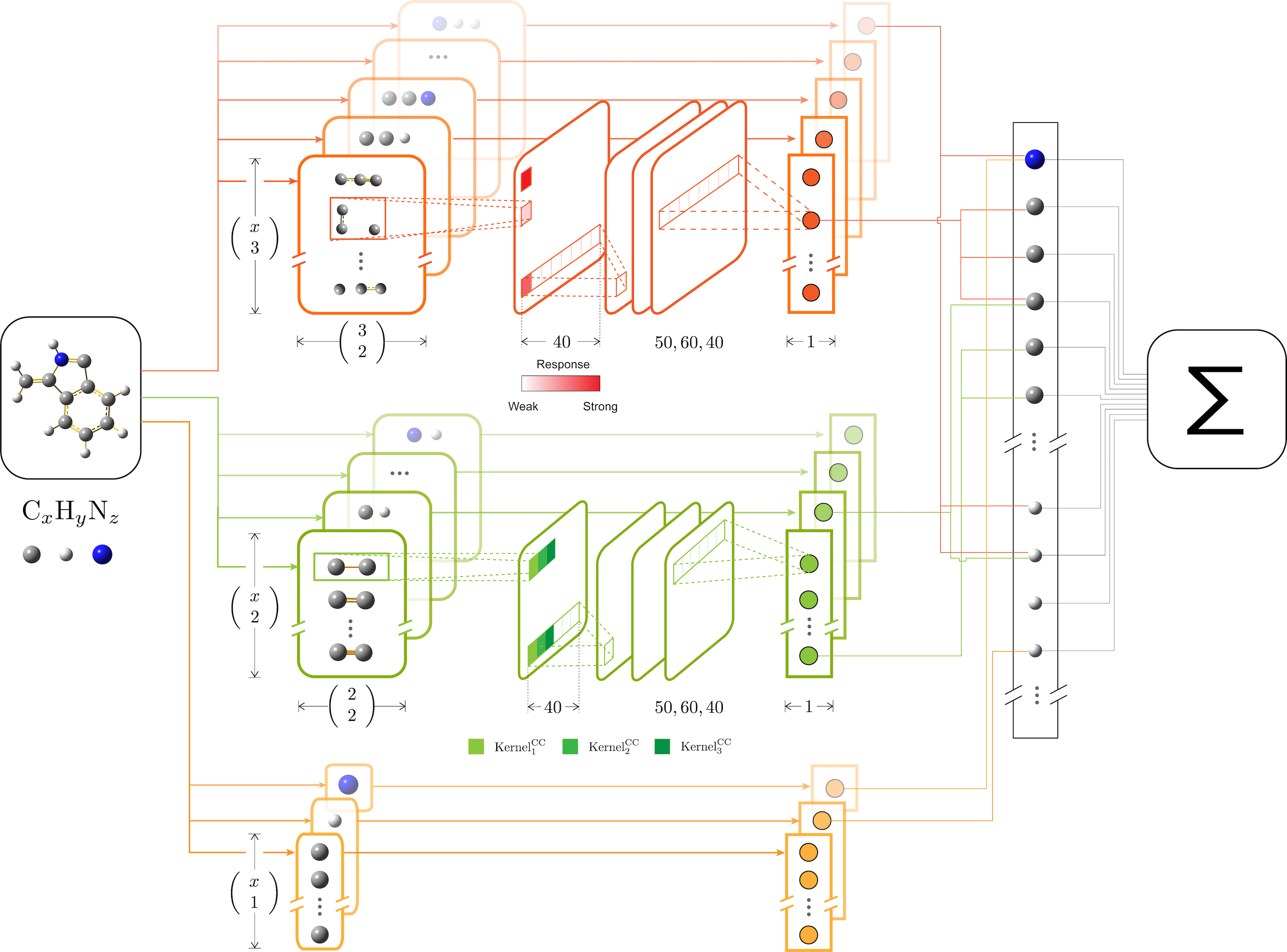
kCON is a scalable and transferable deep learning framework for chemistry with the ability to provide insight into atomistic structures of varying stoichiometry from small and scrap training sets.
- Author: Xin Chen
- Email: Bismarrck@me.com
- Chen, X., Jørgensen, M. S., Hammer, B., & Li, J. (2017). Understanding atomic chemistry with machine learning. Nature Communications, under review.
All codes are written in Python3.6. Some codes can be executed perfectly under Python2.7.x.
- tensorflow>=1.3
- numpy
- scipy
- jupyter
- matplotlib
- scikit-learn
- ase>=3.12
See this guide for using this package.
In order to pull these dataset files from github, Git LFS should be installed.
There some built-in dataset, including:
- QM7
- GDB-9
- Napthalene20k
- Quinoline (PBE, DFTB)
- Anatase Titanium Dioxide (DFTB)
See this guide for more details about how to build these datasets.
See this guide for more details about how to generate the xyz files of these datasets from raw files.
This project is organized like other projects under tensorflow/models.
inference.py: for the detailed implementation of the kCON model.kcnn.py: for constructing kCON model and building the loss function for CPU/single-GPU training.transformer.py: for transformingase.Atomsto input features of the model.save_model.py: for freezing and exporting trained model topbfiles.
train.py: the CPU/single-GPU version of training.multi_gpu_train.py: for training on a single node with multiple GPUs.distributed_train.py: for training on distributed systems. Not implemented yet.evaludation.py: for continuously evaluating the training performance.
database.py: for parsing xyz files and managing the sqlite database files generated byase.db.pipeline.py: for reading TensorFlow Records files to build input pipeline for training and continuous evaluation.tf.contrib.data.DatasetAPI is utilized.build_dataset.py: for transforming raw xyz files to ASE databases and TFRecords.
predictor.py: for making predictions using trained models.- Currently broken
calculator.py: anase.calculator.Calculatorwrapper ofKcnnPredictor.- Currently broken
constants.py: global constants are defined in this module.utils.py: some helper functions are defined in this module.
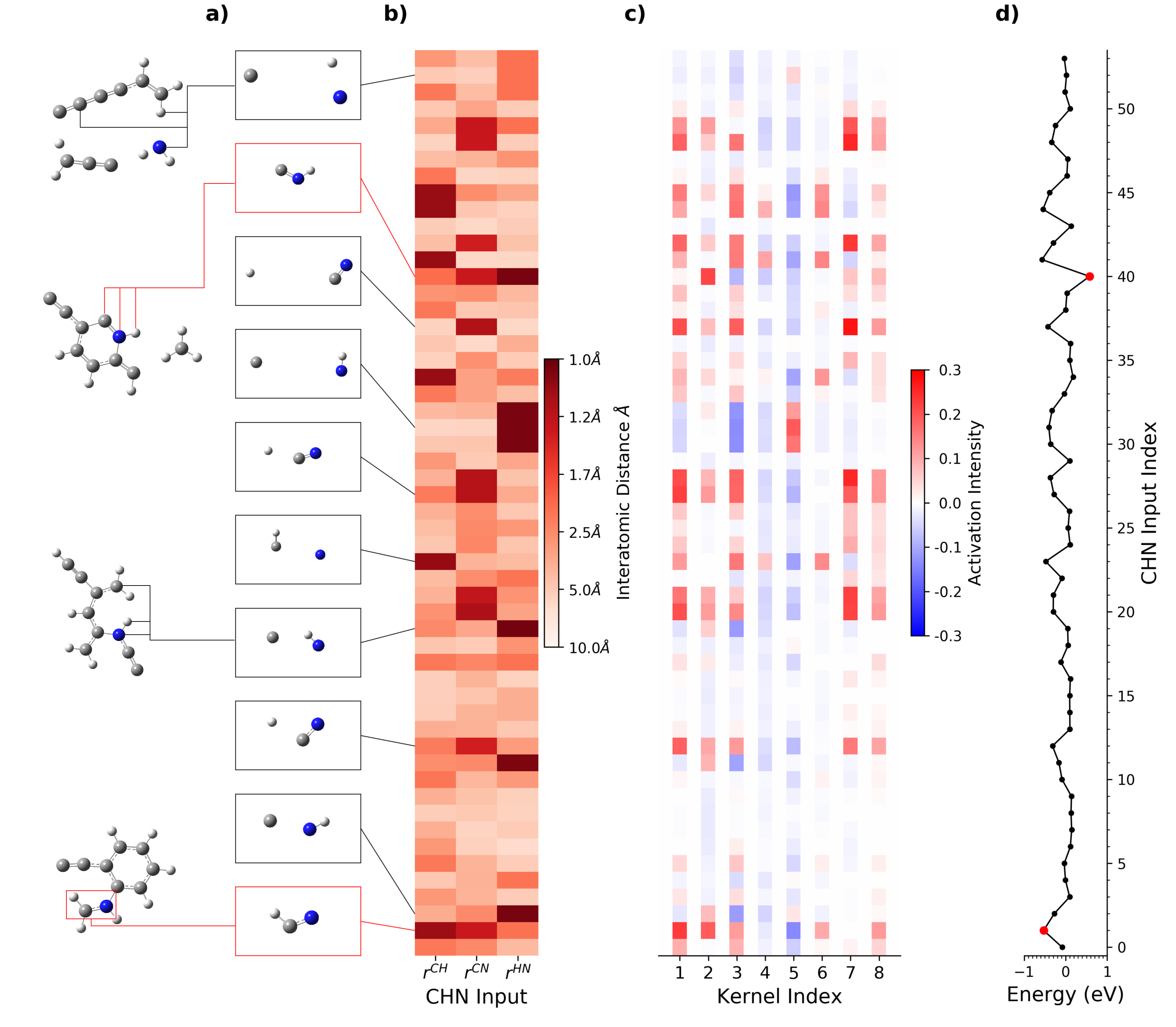
One of the advantage of using convolutional neural network in chemistry is that we can visualize how the network learns chemical patterns. Chemical patterns are somehow very similar to the low-level image features like lines or circles. The traditional CNN visualization methods can be applied directly. Here is a demo of kCON applying on the quinoline PBE dataset.
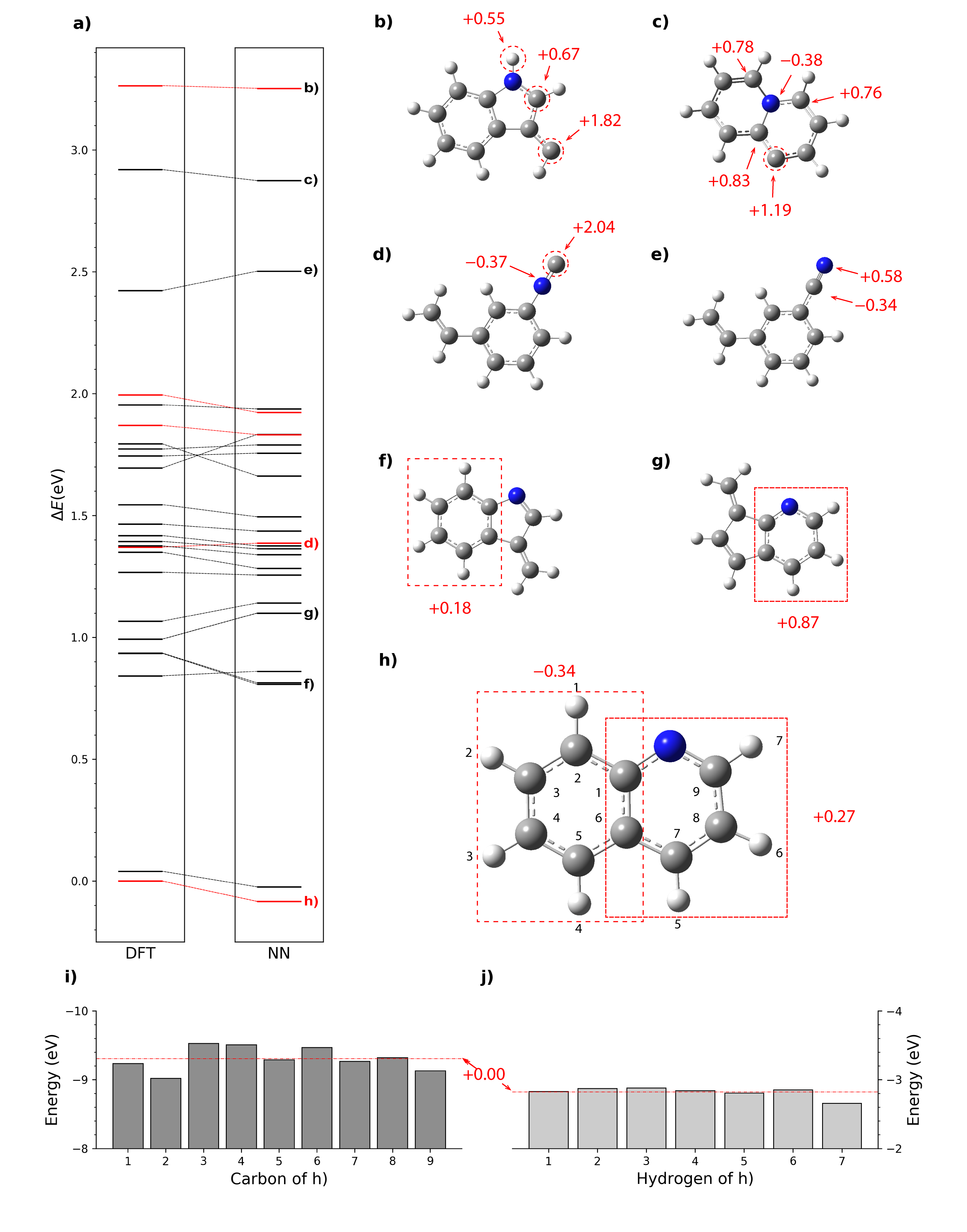
The concept of artificial neural network derived atomic contributions to total energies was first proposed in 2007 by Behler et al. Despite that many later machine learning models include the concept of atomic energy, very little work has been done on interpreting the chemical meaning of these contributions and utilizing them in chemical applications. We did some qualitative and statistic analysis of the atomic energies learned from kCON and successfully proved that these atomic energies can perfectly agree with our chemical intuitions from valence-bond theory, thus providing us with a new approach to understand the local stabilities of atoms and molecular moieties.
Here is an example of using atomic energies to study the stability of molecules.
a) Configurational DFT and Neural Network (NN) energy spectra of Quinoline within 3.5 eV of the global minimum. The DFT energy of the global minimum is the reference energy. Isomers labeled red are included in the test dataset. i) and j) are energies of C and H atoms in the global minimum, and their averages are used as reference for other atomic energies, as shown in h). b), c), d), e), f) and g) are examples of analysis based on relative atomic energies.