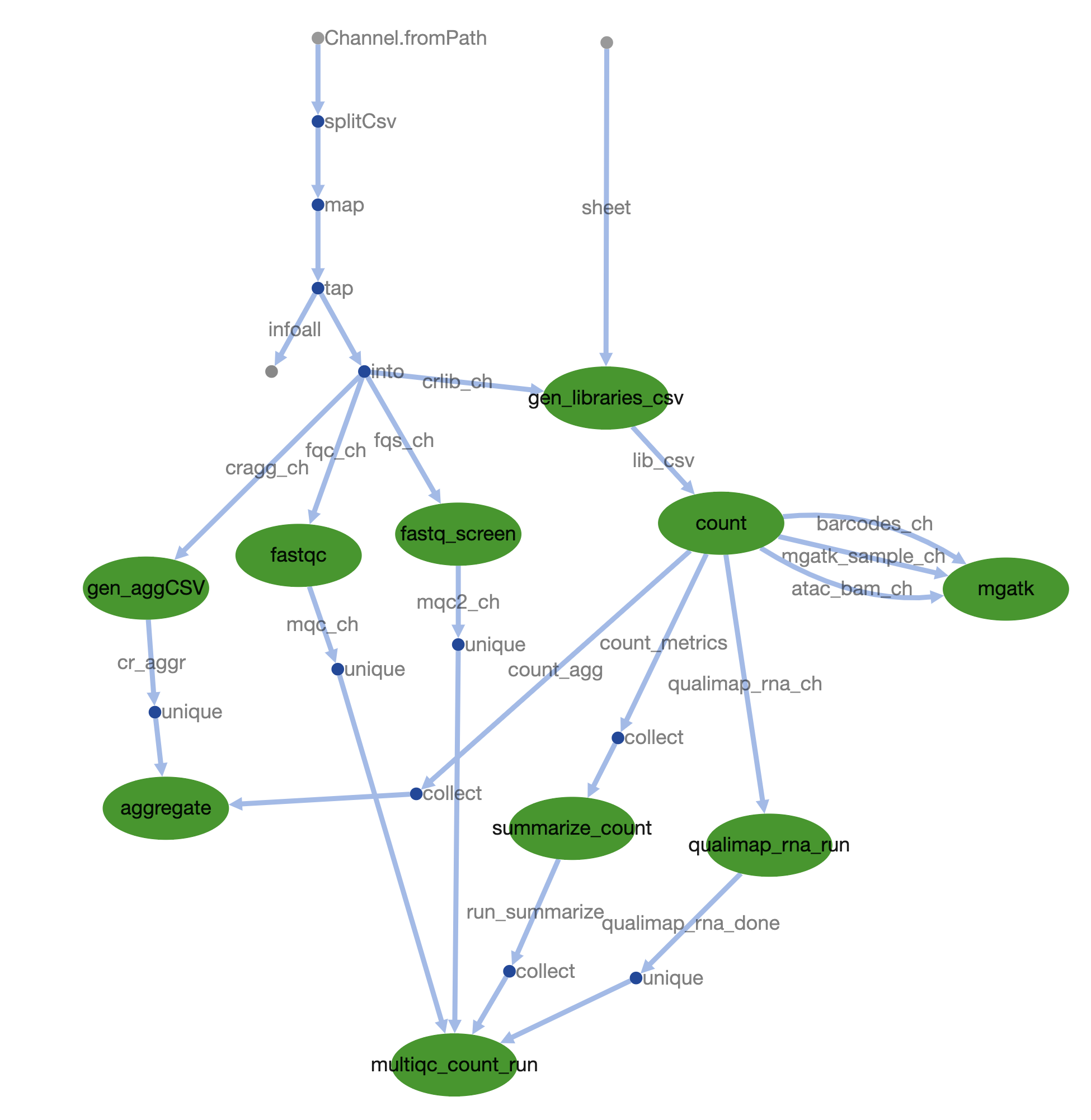
Nextflow pipeline for processing 10x MultiOme datasets with CellRanger-arc and measuring mitochondrial heteroplasmy from ATAC-seq.
-
Analyze 10x multiome (RNA + ATAC) in one pipeline.
-
Starting point are fastq files
| Software | Version |
|---|---|
| nextflow | v.21.10.6 |
| multiqc | v.1.14 |
| mgatk | v.0.6.6 |
| qualimap | v.2.2.1 |
| fastqc | v.0.11.8 |
| fastq_screen | v.0.14.1 |
| cellranger-arc | v.2.0.1 |
| bwa | v.0.7.15 |
| java | min v.1.8 |
Cell Ranger arc was downloaded from 10x (https://support.10xgenomics.com/single-cell-multiome-atac-gex/software/downloads/latest).
You will need to change paths to the software and genomes in the nextflow config file.
Set up a working directory, where you store fastq files, sample table and nextflow config:
- Sample table written in
.csv
with formatting like:
Sample_ID,Sample_Project,Sample_Species,Sample_Lib,Sample_Pair
Sample2,ProjectID,mouse,rna,2
Sample2,ProjectID,mouse,atac,2
Sample1,ProjectID,mouse,rna,1
Sample1,ProjectID,mouse,atac,1
For NUMTs masked genome, it has to be specified in sample table as below:
Sample_ID,Sample_Project,Sample_Species,Sample_Lib,Sample_Pair
Sample2,ProjectID,mouse_NUMTs_masked,rna,2
Sample2,ProjectID,mouse_NUMTs_masked,atac,2
Sample1,ProjectID,mouse_NUMTs_masked,rna,1
Sample1,ProjectID,mouse_NUMTs_masked,atac,1
- Organise directory with folder
fastqand subfoldersatacandrna(fastq.gzfiles can be soft linked):
fastq --|---rna ---|--Sample2_S1_L001_I1_001.fastq.gz
| |--Sample2_S1_L001_I2_001.fastq.gz
| |--Sample2_S1_L001_R1_001.fastq.gz
| |--Sample2_S1_L001_R2_001.fastq.gz
|
|---atac---|--Sample2_S1_L001_I1_001.fastq.gz
|--Sample2_S1_L001_R1_001.fastq.gz
|--Sample2_S1_L001_R2_001.fastq.gz
|--Sample2_S1_L001_R3_001.fastq.gz
- Create nextflow config file using template nextflow.config, and modify directories and paths to genomes and software.
This is already done for Mouse genome in directories (also unmasked genome for Human too):
/suffolk/WorkGenomicsE/mn367/Genomes/CellRanger_Genomes/refdata-cellranger-arc-mm10-2020-A-2.0.0
/suffolk/WorkGenomicsE/mn367/Genomes/CellRanger_Genomes/refdata-cellranger-arc-mm10-2020-A-2.0.0_MT_masked
/suffolk/WorkGenomicsE/mn367/Genomes/CellRanger_Genomes/refdata-cellranger-arc-GRCh38-2020-A-2.0.0
For NUMTs masking of the genome mm10.full.backlist.bed was downloaded from https://github.com/caleblareau/mgatk/wiki/Increasing-coverage-from-10x-processing.
Mask genome using script: mask_NUMTs_genome.sh.
Once the masked fasta is ready, use script CR_mkref.sh to make new CellRanger-arc genome. The config file was required to proceed with making new genome.
Run nextflow on MBU cluster:
change directory to project dir, where fastq files are stored (or linked).
Before running the pipeline, change software paths, e.g. MultiQC, FastQ-screen, qualimap, CellRanger-arc, mgatk etc.
#module load nextflow/22
module load anaconda
module load qualimap
screen -S <session_name>
NXF_VER=21.10.6 nextflow run nf_pipeline_multiome.nf -config nextflow.config -resume