REDCapTidieR
The {REDCapTidieR} package provides an elegant way to import data from a REDCap database into an R environment. It builds upon the {REDCapR} package to query the REDCap API and then transforms the returned data into a set of tidy tibbles.
{REDCapTidieR} is especially useful for dealing with complex databases that are longitudinal or include repeated instruments or both.
Installation
You can install the development version of REDCapTidieR like so:
devtools::install_github("CHOP-CGTDataOps/REDCapTidieR")Getting Started
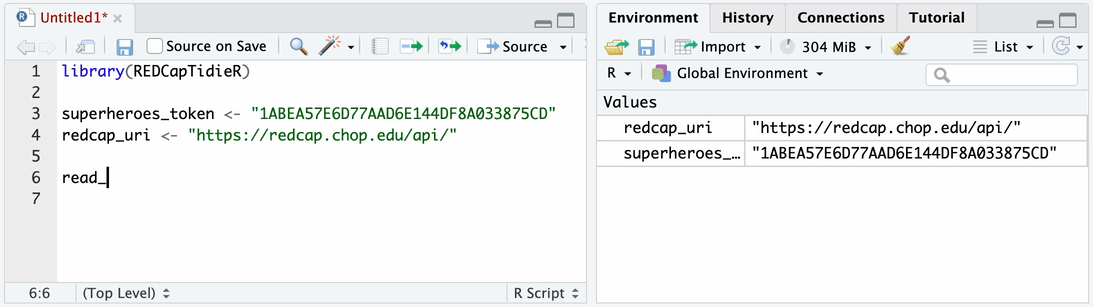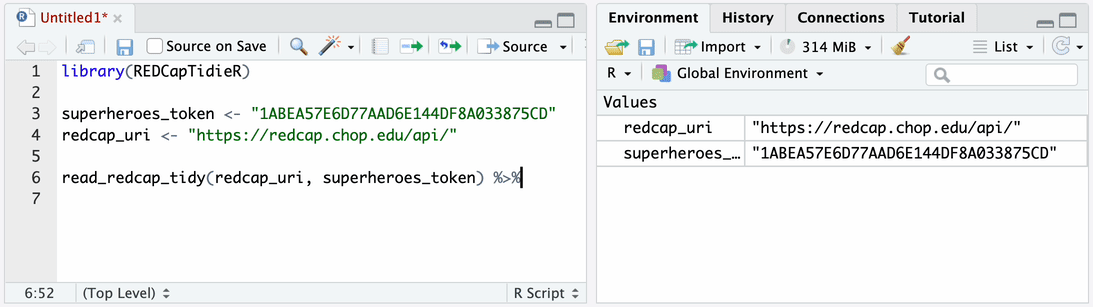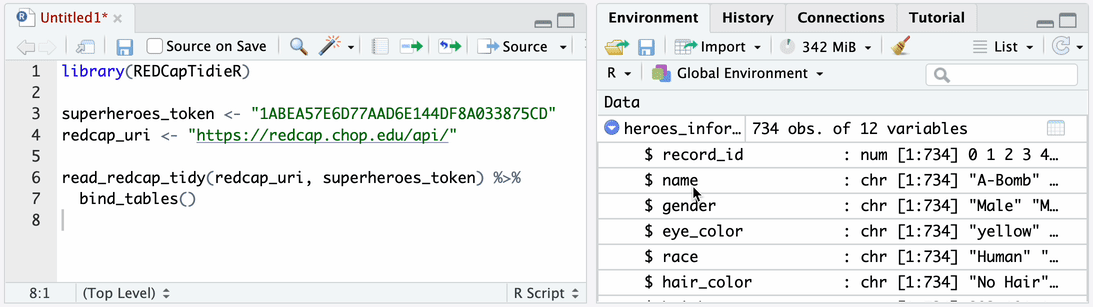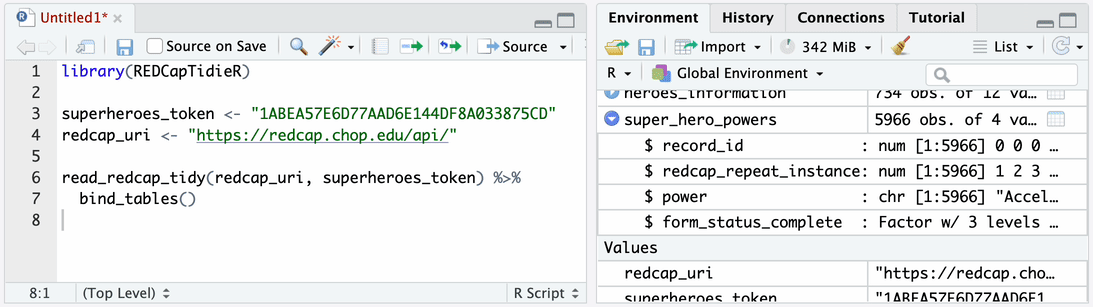
Use read_redcap_tidy() together with bind_tables() to import data
from all instruments into your environment.
Read vignette("getting-started") to learn more.
Collaboration
We invite you to give feedback and collaborate with us! If you are familiar with GitHub and R packages, please feel free to submit a pull request. Please do let us know if {REDCapTidieR} fails for whatever reason with your database and submit a bug report by creating a GitHub issue.
Please note that this project is released with a Contributor Code of Conduct. By participating you agree to abide by its terms.
We’d like to thank the following folks for their advice and code contributions: Will Beasley and Paul Wildenhain.
Funding
This package was developed by the Children’s Hospital of Philadelphia Cell and Gene Therapy DataOps Team to support the needs of the Cellular Therapy and Transplant Section. The development was funded using the following sources:
-
Stephan Kadauke Start-up funds. Stephan Kadauke, PI, CHOP, 2018-2024
-
CHOP-based GMP cell manufacturing (MFG) for CAR T clinical trials. Stephan Grupp, PI; Stephan Kadauke, co-PI, CHOP, 2020-2022