Semiautomated layout of RNA tertiary structure diagrams
(C) Rhiju Das, Stanford University, 2017-2019
Please e-mail questions to ribokit.info@gmail.com
Ribodraw has been tested on MATLAB R2016a and MATLAB 2017b. Some functionality makes use of the Mapping Toolbox and maybe other Toolboxes.
Download RiboKit either by cloning this repository:
git clone https://github.com/RiboKit/RiboDraw.git
or check for the latest release at:
https://github.com/RiboKit/RiboDraw/releases
Then add the RiboDraw/scripts/ directory to your MATLAB path using the command pathtool or Set path... from the menu. Make sure to use the option to include all subdirectories.
You'll need Rosetta's rna_motif executable to extract information on sequence, secondary structure, noncanonical pairs, motifs, etc. Grab the latest release at RosettaCommons and follow the installation instructions at RosettaCommons. Alternatively, you can run a rna_motif job via the rna_info ROSIE server.
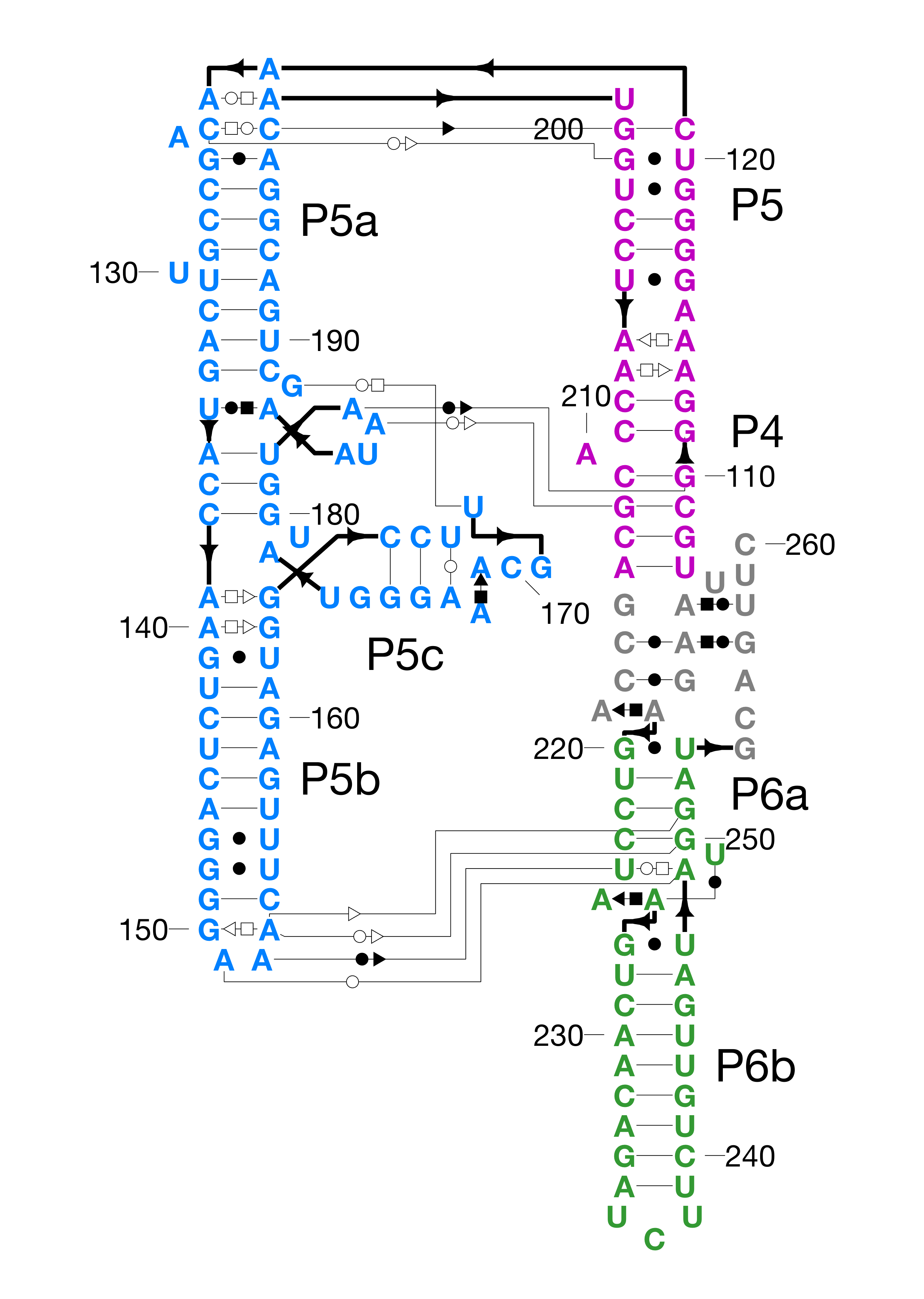
Follow the Tutorial, which teaches you how to make a nice layout of the P4-P6 domain of the Tetrahymena group I self-splicing intron:
It takes about 30 minutes (or more, depending on how much you want to refine the drawing).
A brief account of key steps in the tutorial is available on YouTube; click the thumbnail below!
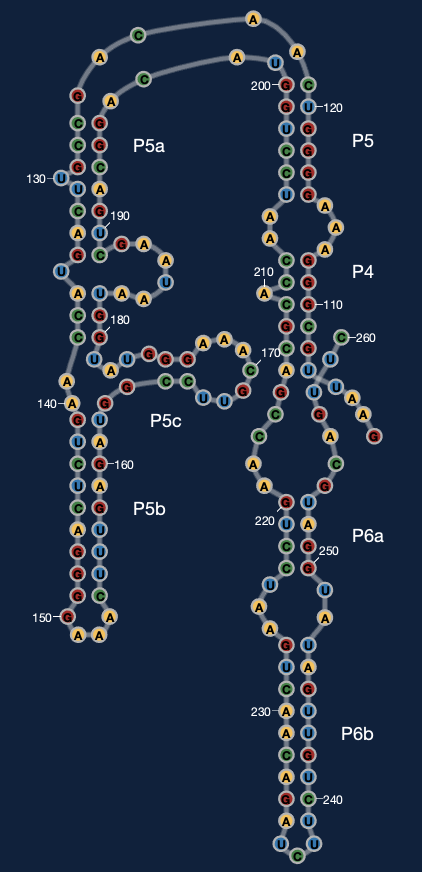
It is also possible to use RiboDraw to generate customLayout's for Eterna puzzles -- this is a preliminary solution for conveying 3D information and resolving overlaps in ribosome-scale problems in Eterna. Here's an example for P4-P6,
available in this drawing, and how this looks inside Eterna at the Eterna P4-P6 development puzzle. Special information on using Ribodraw to prepare Eterna puzzles is available at eterna_tutorial.md.
Documentation of all MATLAB source code is compiled into HTML format at docs. It doesn't directly display in GitHub (yet), but if you open this README.md on your local machine, double click on scripts/docs/menu.html
The core format for RiboDraw 'drawing' files was originally JSON-like to allow for eventual reading and writing with versions of RiboDraw in other languages or other kinds of software, but MATLAB's tools to read/write JSON are currently very slow. The fields are described here. The format can therefore also be saved to .mat MATLAB workspace files, which are very fast to read/write. Drawings can be exported to .png, .jpg, .ps, and .pdf format for manipulation with other software.
- Ribodraw is Open Source. Want to improve RiboKit's MATLAB interface? Port to JavaScript? Check out our Issues page on GitHub. Feel free to fork and make pull requests.
- Running unit tests Go to
RiboDraw/unittestsand typeruntests. Add your own tests to RiboDrawTest.m - Its easy to generate HTML docs We are currently using M2HTML to quickly generate docs for MATLAB scripts. . Download it here, and run the
generate_ribodraw_docscommand in MATLAB to update docs.