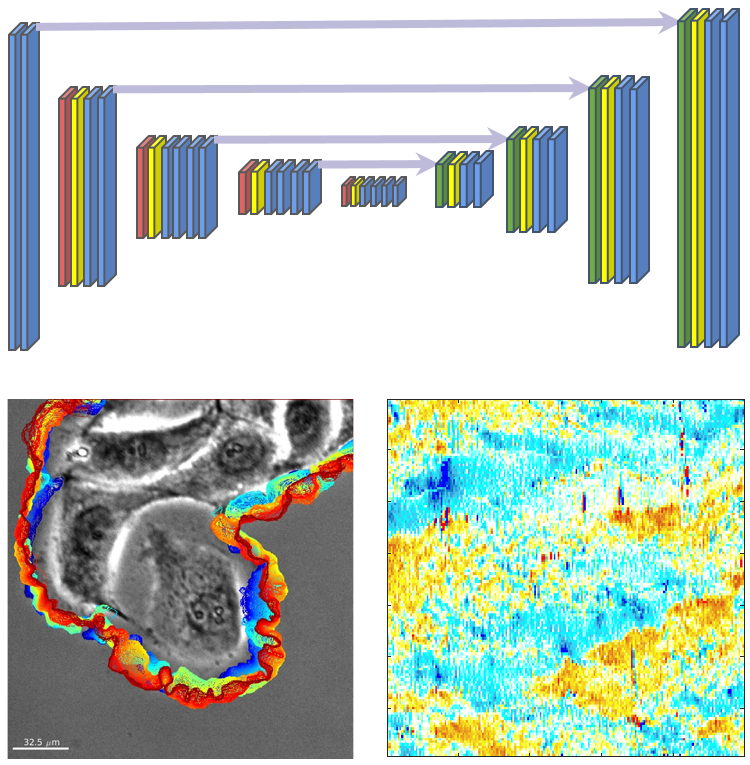
A deep learning-based segmentation pipeline for profiling cellular morphodynamics using multiple types of live cell microscopy
by Junbong Jang, Chuangqi Wang, Xitong Zhang, Hee June Choi, Xiang Pan, Bolun Lin, Yudong Yu, Carly Whittle, Madison Ryan, Yenyu Chen, Kwonmoo Lee
To learn more about our pipeline (MARS-Net), please read the paper
You can quickly segment one of our live cell movie in this demo (Estimated Time: 12 minutes)
This demo allows users to see the segmentation performance of MARS-Net and U-Net which are already trained on our live cell movies. To test MARS-Net pipeline from the scratch in a user's local machine, the user needs to satisfy software requirements and train the models before segmenting movies.
MARS-Net pipeline has been tested on Windows 10 and Ubuntu 16.04, 18.04 and 20.04 with anaconda v4.5.11 and Python v3.6
- For evaluation and visualization, we used
- MATLAB 2019b
- To read npy files generated from Python, NPY Reader
- To calculate F1, precision and recall, Correspondence Algorithm developed by University of California Berkeley Segmentation Benchmark
- For Morphodynamics Profiling, we used
- Windowing and Protrusion package developed by Gaudenz Danuser lab (UT Southwestern) for Morphoydynamics profiling.
- For training the deep learning models and segmenting live cell movies, we used
- Tensorflow v2.3 on RTX Titan GPU with CUDA 10.1
- Tensorflow v1.15 on GTX 1080Ti GPU with CUDA 10.0
- Other Python dependencies in the Anaconda environment are specified in environment.yml
The pipeline consists of label tool, segmentation modeling, and morphodynamics profiling.
Installation Time can vary based on user's download speed (Estimated Time: 1 hour)
- Download MARS-Net pipeline from Github repository and install its software requirements.
- Setup Anaconda environment
- In Linux OS
-
conda env create --name marsnet --file environment_linux.yml
-
- In Windows 10
-
conda env create --name marsnet --file environment_windows.yml
-
-
conda activate marsnet
-
pip install tensorflow-addons
- In Linux OS
- Before running the pipeline, please specify the following parameters in UserParams.py
- strategy_type (The type of deep learning model. e.g. specialist_unet, or generalist_VGG19_dropout)
- dataset_folders (location where your images and mask are stored)
- img_type (type of image. default is .png)
- img_folders (list of image folder names)
- mask_folders (list of mask folder names)
- frame_list (list of training frames. e.g. [1,2,6,10,22,34])
- dataset_names (list of dataset folder names)
- model_names (list of the model names, necessary since multiple models are created from cross validation)
- REPEAT_MAX (Max number of times to repeat cross validation. e.g. 1 or 5)
- Other parameters can be ignored
- Store movie datasets in the directories specified in UserParams.py.
Each folder must contain images and their corresponding masks of one movie.
- phase contrast movie with its labeled mask is in repository's assets folder.
- single-microscopy-type U-Net and multiple-microscopy-type VGG19D-U-Net trained on 2 frames per movie in leave-one-movie-out cross validation
Facilitates labelling raw images semi-automatically and it is located in label_tool folder.
- Specify the location of image files to label in user_params.py
- set dataset name to a_dataset variable and image folder path to img_root_path
- To determine the optimal hysteresis thresholding for canny detector and kernel size for blurring,
- python explore_edge_extraction_user_params.py
- Compare results in generated_explore_edge folder
- Then, set the best hyper parameters in user_params.py
- canny_std_multiplier and denoise_kernel_size,
- To extract edge,
-
python extract_edge.py
- The generated edge images are saved in generated_edge folder
-
- Manually fix the generated edge images
- Post processing to fill the edge images
- python segment_edge.py
- segment_edge.py script will ask for how many backgrounds to fill in your image and one pair of (x,y) coordinate in each background area.
- The post processed results are saved in generated_segmentation folder
- please move these labeled images into the assets folder to train the model.
This section is for training deep learning models from scratch and segmenting the live cell movies
- Put your live cell movies into the assets folder. Our pipeline assumes leave-one-movie-out cross validation so please provide multiple movies.
- To crop patches
-
python crop/crop_augment_split.py
-
- To train on the cropped patches
-
python models/train_mars.py
-
- To segment live cell movies
-
python models/prediction.py
-
This section is for replicating our evaluation results including bar graphs, line graphs, and violin plots.
- Edit parameters in evaluation/GlobalConfig.m
- prediction_path_list
- display_names
- frame_list
- root_path
- img_root_path
- Download NPY Reader and add the folder to MATLAB path.
- Download Correspondence Algorithm and add the folder to MATLAB path.
- Open MATLAB to run the scripts.
- If a user prefers to run them in the terminal, type the following command after replacing ##### with the actual script name
-
matlab -nodisplay -nosplash -nodesktop -r "run('#####.m');exit;"
-
- If a user prefers to run them in the terminal, type the following command after replacing ##### with the actual script name
- Before visualizing the evaluated results, calculate F1, precision and recall from the segmented movies, run
- evaluation/evaluation_f1/run_overlap_mask_prediction.m
- To draw bar graphs and line graphs across different training frames, run
- evaluation/evaluation_f1/visualize_across_frames_datasets.m
- To draw edge evolution, run
- evaluation/evaluation_f1/run_overlap_compare.m
- To draw violin plot, run
- evaluation/violin_compare.m
Unlike MATLAB code above, learning curves and bubble plots are drawn using Python
- To draw learning curve
-
python evaluation/draw_learning_curve.py
-
- To draw bubble plot, open the following jupyter notebook and run all cells
- evaluation/bubble_training_curve.ipynb
To draw activation maps and replicate SEG-Grad-CAM results
-
python SegGradCAM/main.py
- For single cell cropping of the segmented movie
- Post process the segmented images to binarize them and fill small holes in MATLAB
- process_predicted.m
- Rotate and crop the image in a way that a single cell is within the cropped window
-
python img_proc/rotate_crop_img.py
-
- Post process the segmented images to binarize them and fill small holes in MATLAB
- Download Windowing and Protrusion package
- In MATLAB, add directory and subdirectory of the downloaded package to the MATLAB path
- Type movieSelectorGUI in the MATLAB command window
- Create new movie and perform windowing on the segmented movie from the previous step.
- For details, refer to Windowing and Protrusion package Github page.