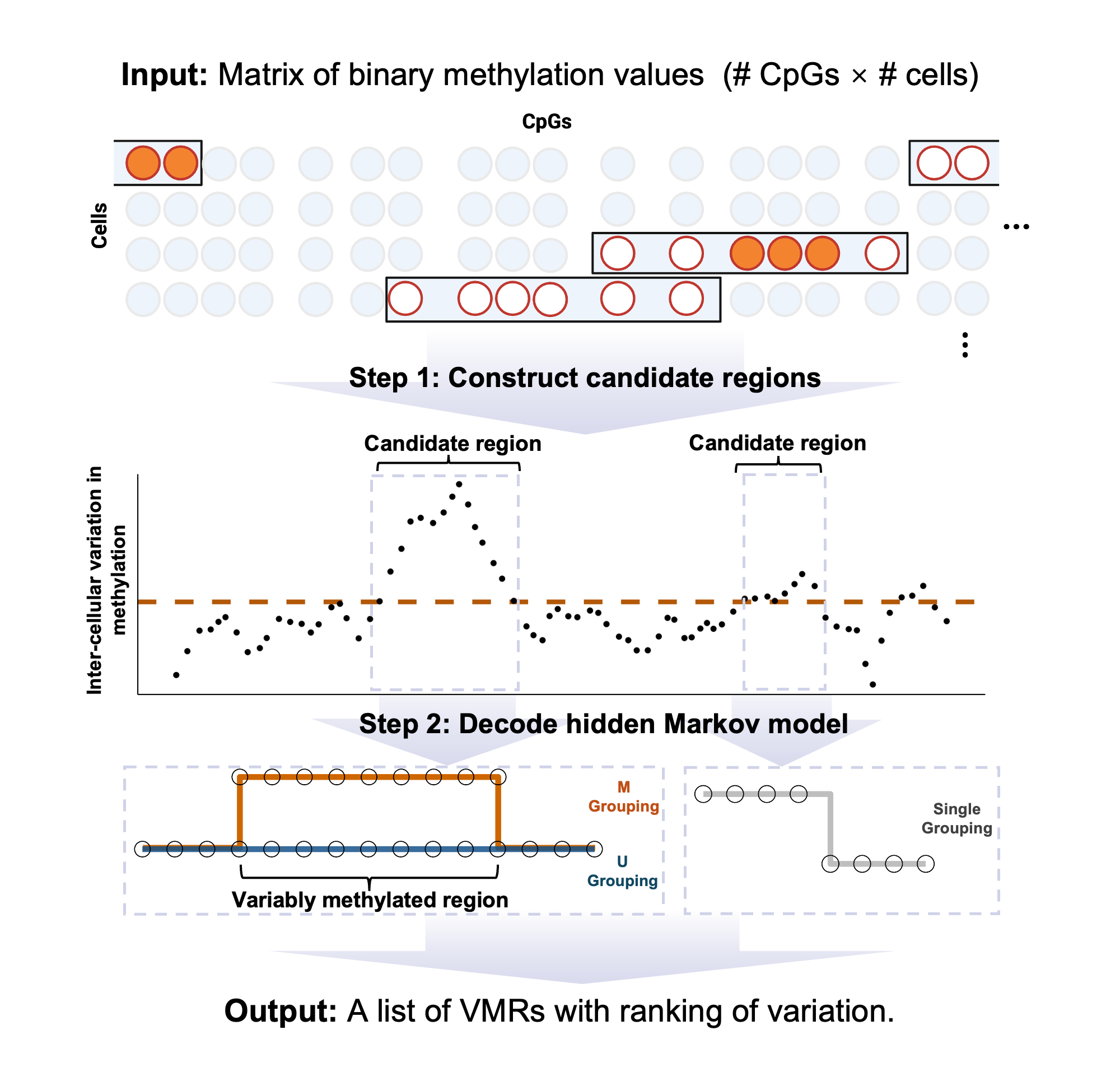
The R package vmrseq is a novel computational tool developed for pinpointing variably methylated regions (VMRs) in scBS-seq data without prior knowledge on size or location. High-throughput single-cell measurements of DNA methylation allows studying inter-cellular epigenetic heterogeneity, but this task faces the challenges of sparsity and noise. vmrseq overcomes these challenges and identifies variably methylated regions accurately and robustly.
You can install the development version of vmrseq in R from
GitHub with:
# install.packages("devtools")
devtools::install_github("nshen7/vmrseq")- An online vignette of 'Get Started' on the
vmrseqpackage can be found at https://rpubs.com/nshen7/vmrseq-vignette. - An example workflow on scBS-seq data analysis using
vmrseqcan be found at https://github.com/nshen7/vmrseq-workflow-vignette.
We provide a Docker image for robust setup and use of this package. The Docker image includes all necessary dependencies for the package and vignettes. To pull the Docker image from Docker Hub, use the following command in bash:
docker pull nshen7/vmrseq-bioc-3.19:latestNing Shen and Keegan Korthauer. 2023. “Vmrseq: Probabilistic Modeling of Single-Cell Methylation Heterogeneity.” bioRxiv. https://doi.org/10.1101/2023.11.20.567911.