Ying Zhang 26 December, 2022
This repository contains documentation for the phenotype, sleep monitoring (e.g. polysomnography, polygraphy, actigraphy) and survey questionnaire data harmonized by the National Sleep Research Resource (NSRR), including harmonization standards, rationale, and script for recreating the harmonized data.
| file | description |
|---|---|
non-sleep-phenotype/ |
A directory containing documentations for harmonized non-sleep covariates. |
sleep-monitoring/ |
A directory containing documentations and script for creating harmonized sleep monitoring data. |
sleep-questionnaire/ |
A directory containing documentations and script for creating harmonized sleep questionnaire data. |
README.md |
This document. |
README.Rmd |
Source for this document. |
Table of Contents
The objectives for data harmonization is (1) to identify core sleep terms that are semantically similar, (2) to assess the heterogeneity in the study-level and variable-level metadata, (3) to improve comparability of the harmonized terms, and (4) to make harmonized terms inferentially equivalent when possible. We identified three types of data currently shared on the NSRR website that are most likely to be valuable to our users once harmonized - commonly used non-sleep covariates, sleep monitoring data (e.g.,polysomnography/polygraphy) and sleep questionnaire data. We’ve developed domain-specific approach to address the challenges in harmonizing each of these types of data. Harmonized data were created using SAS and saved as a separate data file in each dataset on NSRR website. All SAS scripts are publicly available on github.com/nsrr.
It is worth noting that the current harmonization process is not data-driven but NSRR is planning on assessing how similar harmonized data is across demographically similar datasets.
TOPMed DCC’s harmonized
phenotype
and BioDataCatalyst Common Data
Model
were identified as the harmonization standards for the non-sleep
phenotype data. We have harmonized 9 non-sleep phenotype variables:
age(nsrr_age), sex(nsrr_sex), race(nsrr_race),
ethnicity(nsrr_ethnicity), body mass index(nsrr_bmi), current
smoker(nsrr_current_smoker), ever smoker(nsrr_ever_smoker), resting
systolic(nsrr_bp_systolic) and diastolic blood
pressure(nsrr_bp_diastolic).
The original data format was preserved for numeric variables;
Categorical variables (e.g., sex, race) were recoded as string. All
permissible values for the harmonized categorical variables are
available as JSON files in non-sleep-phenotype/.
For numerical variables, missing values were represented as empty cell value in the date files. For categorical variables, explicit missing codes were used to comply with the TopMED/BioDataCatalyst harmonization standards. Unspecific missing was coded as “not reported” for most categorical variables.
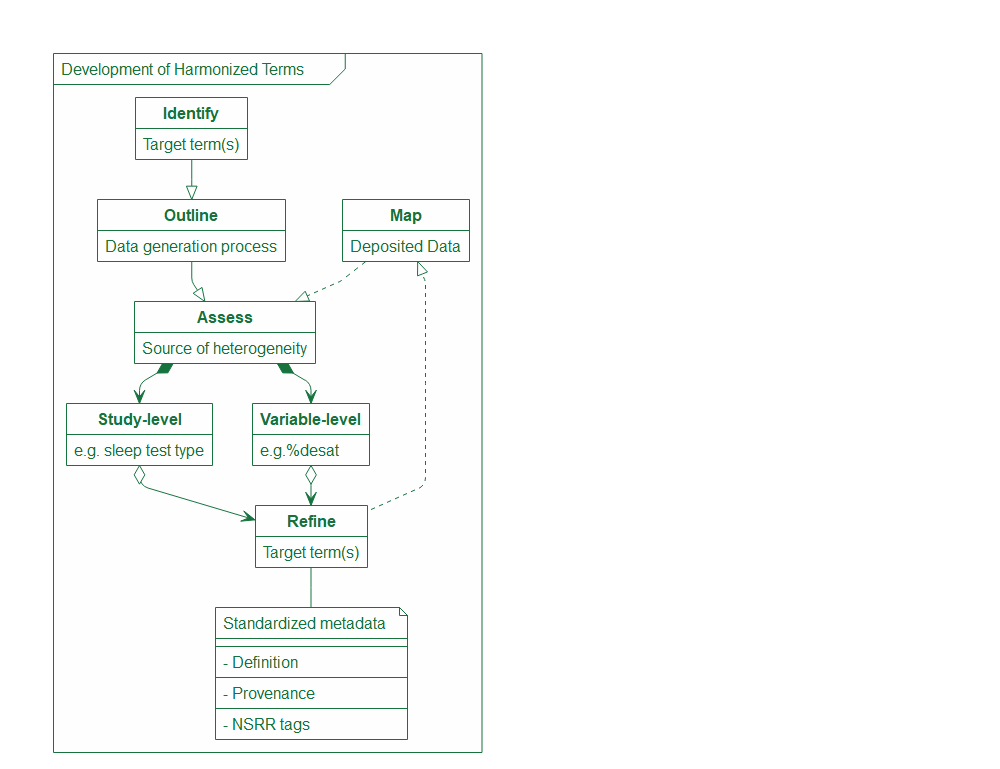
Retrospective polysomnography/Polygraph (PSG) data harmonization focuses on summary PSG data submitted by the data owners, which was either manually scored by scorers or automatically scored by the device. Prospective PSG data harmonization using EDF files can be done by the NSRR NAP pipeline. As illustrated in the process diagram down below, we took an iterative approach when developing harmonized summary PSG terms. We first identified the initial target terms (e.g., AHI3%, AHI4%, total sleep duration, overall arousal index and sleep/wake signal quality flag). Then we mapped out the key steps in the PSG data generation process which tend to be the major sources of heterogeneity. After identifying all the varying components in the study-level and variable-level metadata that greatly affect the comparability of PSG data, we went back to the initial target terms and decided whether (and how) we would need to modify and expand the target terms to accommodate all the heterogeneity. We then mapped the deposited data at the NSRR to the refined target terms and assessed if there were any additional sources of heterogeneity that should be accounted for. We also standardized the metadata for the target terms during the process which includes harmonized term definitions, provenance information (how to document the source data and any processing of the source data), as well as the standardized NSRR tags for “forward-compatible” harmonization.
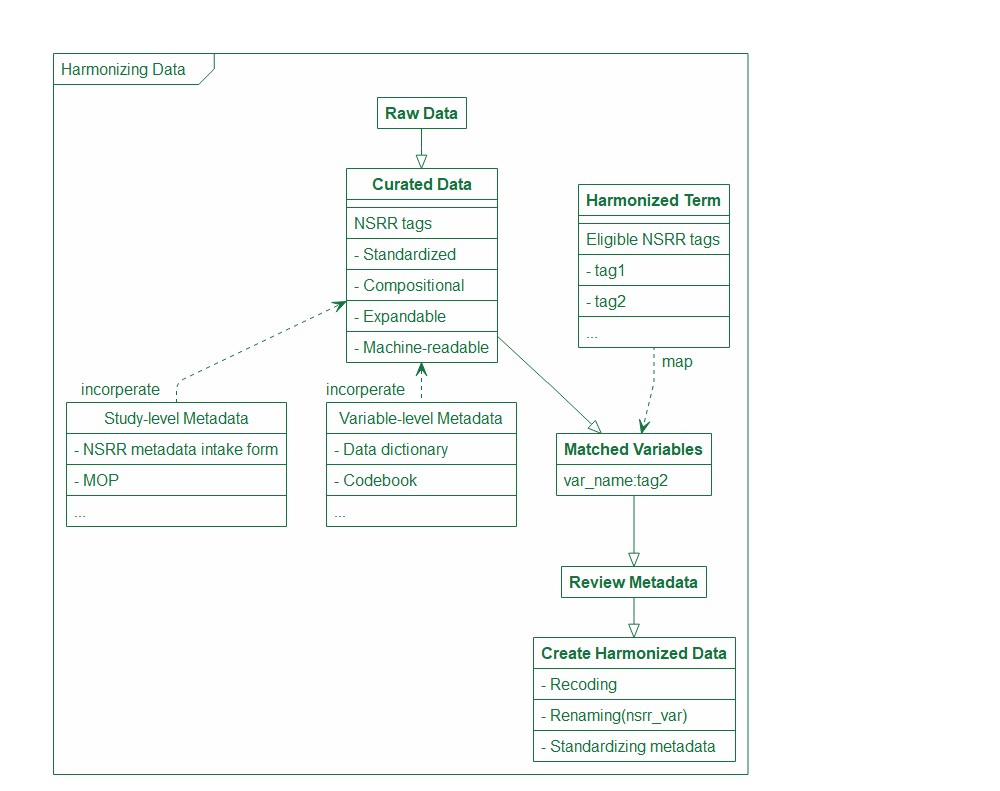
Using the NSRR tags, we are able to leverage data/metadata curation that we do routinely at NSRR to quickly map and create new harmonized data. As shown in the diagram down below, once the raw data is curated and assigned a NSRR tag based on a compositional coding scheme we developed at NSRR, we can simply search if any of the eligible tags listed by the harmonized term(s) metadata is present in the new dataset. Once there is a match, we will conduct an in-depth review of all the associated metadata to make sure the harmonized term(s) are mapped correctly, and create a harmonized version with new variable name and metadata once the mapping is confirmed.
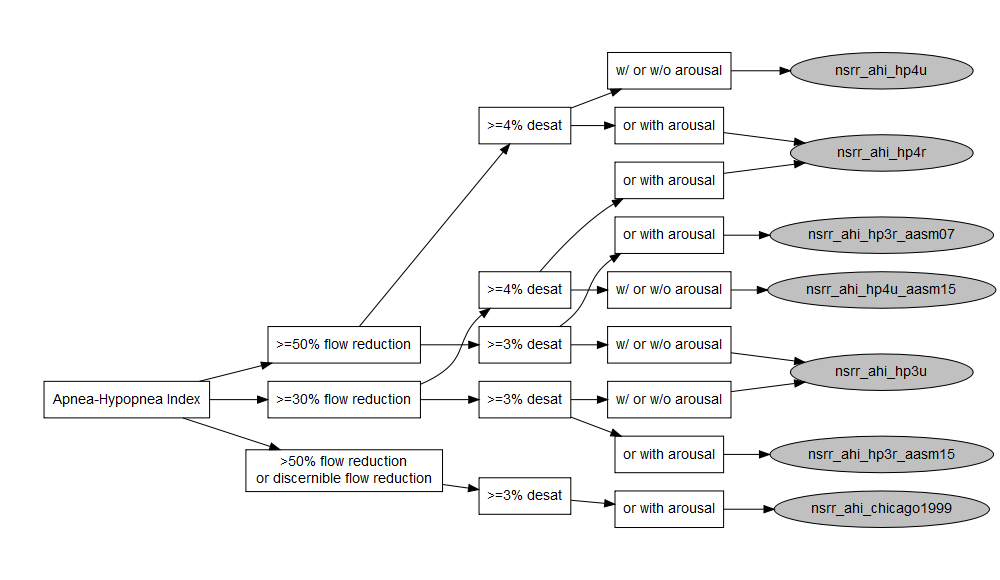
After carefully evaluating the main sources of heterogeneity in PSG data generation and processing across datasets, we’ve identified four varying components in the metadata - sleep test type (i.e., type I or II polysomnography, type III or IV polygraphy/home sleep apnea test), flow reduction, oxygen desaturation, and arousal. Here is a diagram for how commonly referred to as AHI3% and AHI4% can vary in the these categories and create many more permutations.
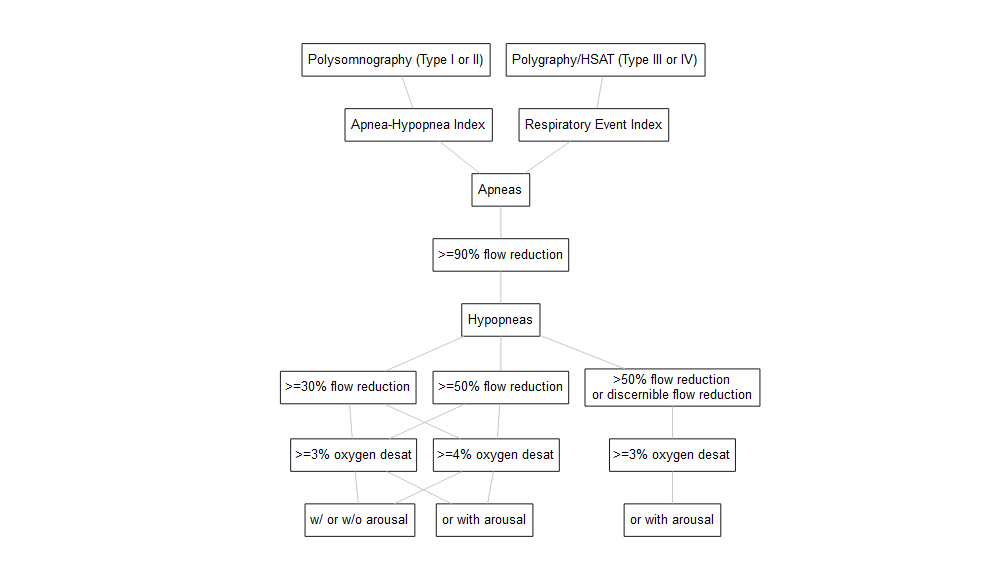 Among the 13
permutations of AHI3% and AHI4% as illustrated in the diagram above,
we’ve mapped a subset of them to the American Academy of Sleep Medicine
(AASM) Clinical Guidelines and consolidated into 10 harmonized
variables - 7 AHIs and 3 REIs.
Among the 13
permutations of AHI3% and AHI4% as illustrated in the diagram above,
we’ve mapped a subset of them to the American Academy of Sleep Medicine
(AASM) Clinical Guidelines and consolidated into 10 harmonized
variables - 7 AHIs and 3 REIs.
| Year | Version | Definition for Hypopnea Events | Event Duration | Reduction Duration Criteria | Consolidated Coding |
|---|---|---|---|---|---|
| 1999 | >50% nasal cannula reduction or discernible nasal cannula reduction with (>=3% desat or arousal) | >=10 sec | chicago1999 | ||
| 2007 | Recommended | >=30% nasal cannula [or alternative sensor] reduction with >=4% desat | >=10 sec | >=90% of the event duration | same as hp4u_aasm15 |
| 2007 | Alternative | >=50% nasal cannula [or alternative sensor] reduction with (>=3% desat or arousal) | >=10 sec | >=90% of the event duration | hp3r_aasm07 |
| 2012 | Recommended | >=30% nasal cannula [or alternative sensor] reduction with (>=3% desat or arousal) | >=10 sec | >=10 sec | same as hp3r_aasm15 |
| 2012 | Alternative | >=30% nasal cannula [or alternative sensor] reduction with >=4% desat | >=10 sec | >=10 sec | same as hp4u_aasm15 |
| 2015 | Recommended | >=30% nasal cannula [or alternative sensor] reduction with (>=3% desat or arousal) | >=10 sec | >=10 sec | hp3r_aasm15 |
| 2015 | Acceptable | >=30% nasal cannula [or alternative sensor] reduction with (>=4% desat) | >=10 sec | >=10 sec | hp4u_aasm15 |
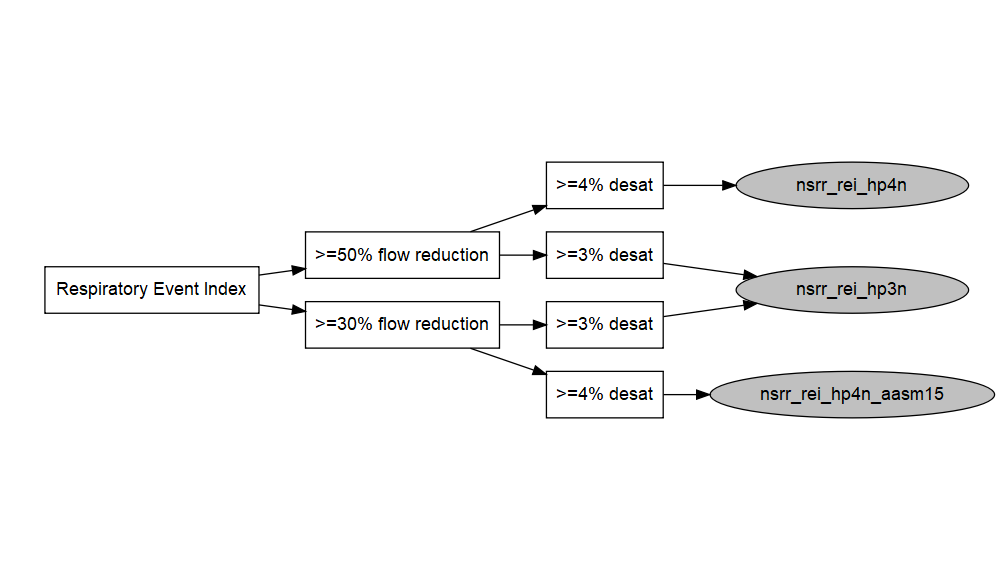
Harmonized AHI/REI terms are shown in grey circles in the diagram above, and can be searched on the NSRR website. Links to the source variables are available in the metadata.
| Harmonized terms | Eligible NSRR tags |
|---|---|
| nsrr_ahi_hp3u | ahi_ap0uhp5x3u_f1t1, ahi_ap0uhp5x3u_f1t2, ahi_ap0uhp3x3u_f1t1, ahi_ap0uhp3x3u_f1t2 |
| nsrr_ahi_hp3r_aasm07 | ahi_ap0uhp5x3r_f1t1, ahi_ap0uhp5x3r_f1t2 |
| nsrr_ahi_hp3r_aasm15 | ahi_ap0uhp3x3r_f1t1, ahi_ap0uhp3x3r_f1t2 |
| nsrr_ahi_hp4u | ahi_ap0uhp5x4u_f1t1, ahi_ap0uhp5x4u_f1t2 |
| nsrr_ahi_hp4u_aasm15 | ahi_ap0uhp3x4u_f1t1, ahi_ap0uhp3x4u_f1t2 |
| nsrr_ahi_hp4r | ahi_ap0uhp5x4r_f1t1,ahi_ap0uhp5x4r_f1t2, ahi_ap0uhp3x4r_f1t1, ahi_ap0uhp3x4r_f1t2 |
| nsrr_ahi_chicago1999 | ahi_ap0uhp5x0u_ap0uhp0x3r_f1t1, ahi_ap0uhp5x0u_ap0uhp0x3r_f1t2 |
| nsrr_rei_hp4n | rei_ap0nhp5x4n_f1t3, rei_ap0nhp5x4n_f1t4 |
| nsrr_rei_hp4n_aasm15 | rei_ap0nhp3x4n_f1t3, rei_ap0nhp3x4n_f1t4 |
| nsrr_rei_hp3n | rei_ap0nhp5x3n_f1t3, rei_ap0nhp5x3n_f1t4, rei_ap0nhp3x3n_f1t3, rei_ap0nhp3x3n_f1t4 |
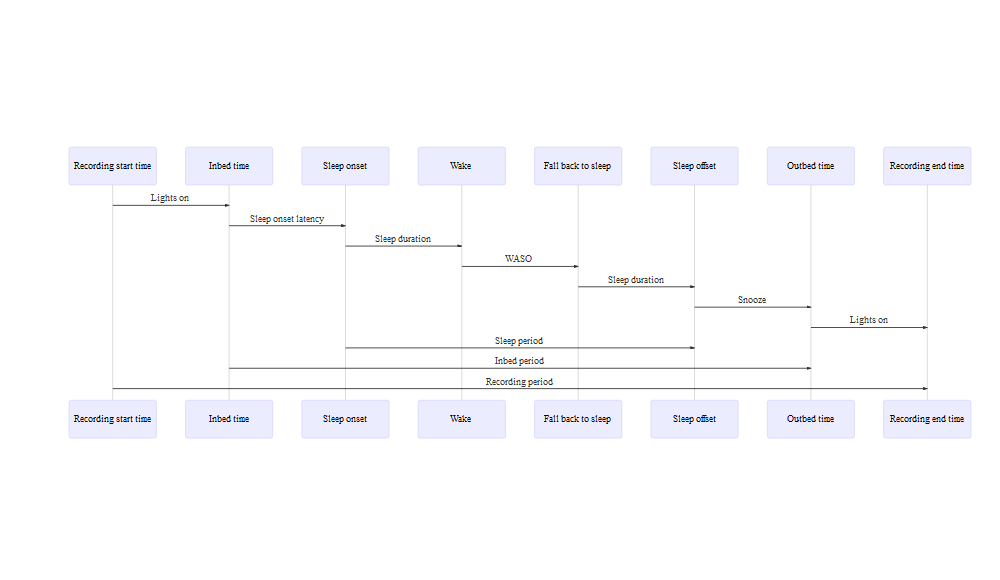
The terms to describe time intervals versus points of time are one of the major sources of confusion and ambiguity in PSG sleep architecture metadata. Depending on the context and usage, “time” might refer to a specific point in time or to an interval between two time points (e.g., study start time versus total sleep time), while “duration” and “period” could both be taken to correspond to intervals, they were often used interchangeably in protocol descriptions and data dictionaries without any indication of whether they referred to intervals between designated time points or to specific intervals when subjects were determined to be awake or asleep. Therefore, we’ve developed a set of standardized terms to address the challenges of “time”, “period”, “duration” being used inconsistently at the NSRR metadata documentation internally.
Total sleep duration (i.e., total sleep time) is defined as the interval
between sleep onset and offset while the subject was asleep, which is an
important measure used in calculating various indices. Among studies
with type I/II polysomnography data, we’ve mapped all total sleep
duration and created a harmonized variable nsrr_ttldursp_f1 in which
f1 refers to the source of data (i.e.,f1=polysomnography/polygraphy,
f2=actigraphy, f3=sleep questionnaire).
| Harmonized terms | Eligible NSRR tags |
|---|---|
| nsrr_ttldursp_f1 | ttldursp_f1t1, ttldursp_f1t2, ttldursp_f1t3 |
Overall arousal index is defined as the total number of arousal divided
by the total sleep duration (i.e., total sleep time). We’ve mapped all
overall arousal index from datasets with type I/II polysomnography and
created a harmonized variable nsrr_phrnumar_f1.
| Harmonized terms | Eligible NSRR tags |
|---|---|
| nsrr_phrnumar_f1 | phrnumar_f1t1, phrnumar_f1t2 |
We’ve selected sleep/wake signal quality flag for it’s commonly
misunderstood and misused by the NSRR users. When the EEG is
insufficient in quality to allow distinction of sleep stages, all sleep
stages were scored using a default of “Stage 2” and no arousal was
scored. Studies and individuals with “sleep/wake only” instead of “full
scoring” for nsrr_flag_spsw should not be used to analyze sleep
architecture or arousals.