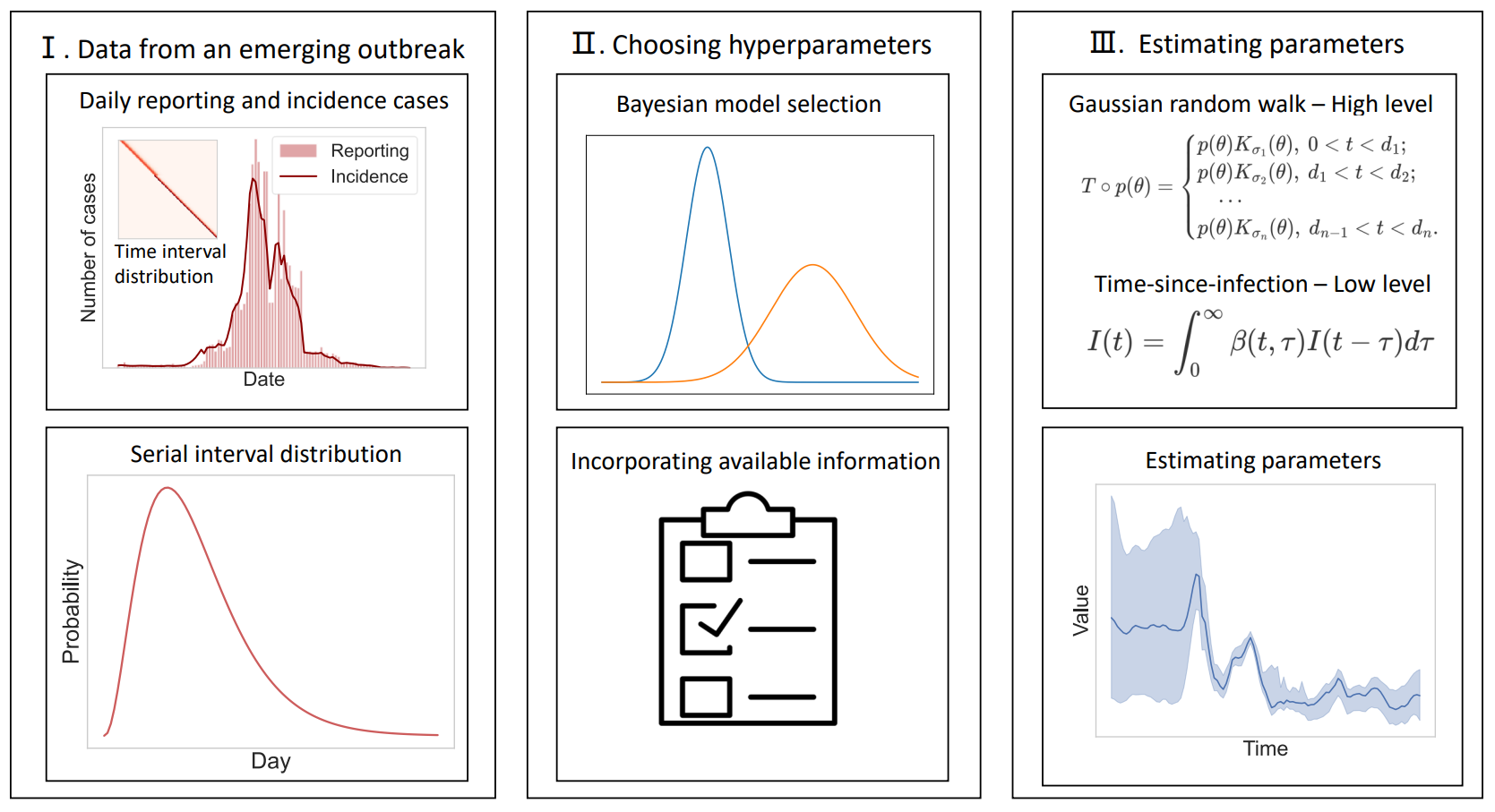
Python implementation of a Bayesian statistical framework for modelling and analysing emerging outbreaks.
The dynamics of an emerging outbreaks is modelled with a hierarchical Bayesian model. On short time scales, the incident was depicted with a renewal process based on Kermack-McKendrick model with key parameters quantifying the effect of mitigation and quarantine, while on long times scales these time-varying parameters were modelled via Gaussian random walk with changing standard deviation. This model allows incorporating of available information about interventions by configuring high-level model, e.g. setting specific structural break points at initialisation of public health measures.
bayesloop==1.5.6
loguru==0.6.0
numpy==1.23.0
optuna==3.0.3
pandas==1.4.3
scipy==1.8.1
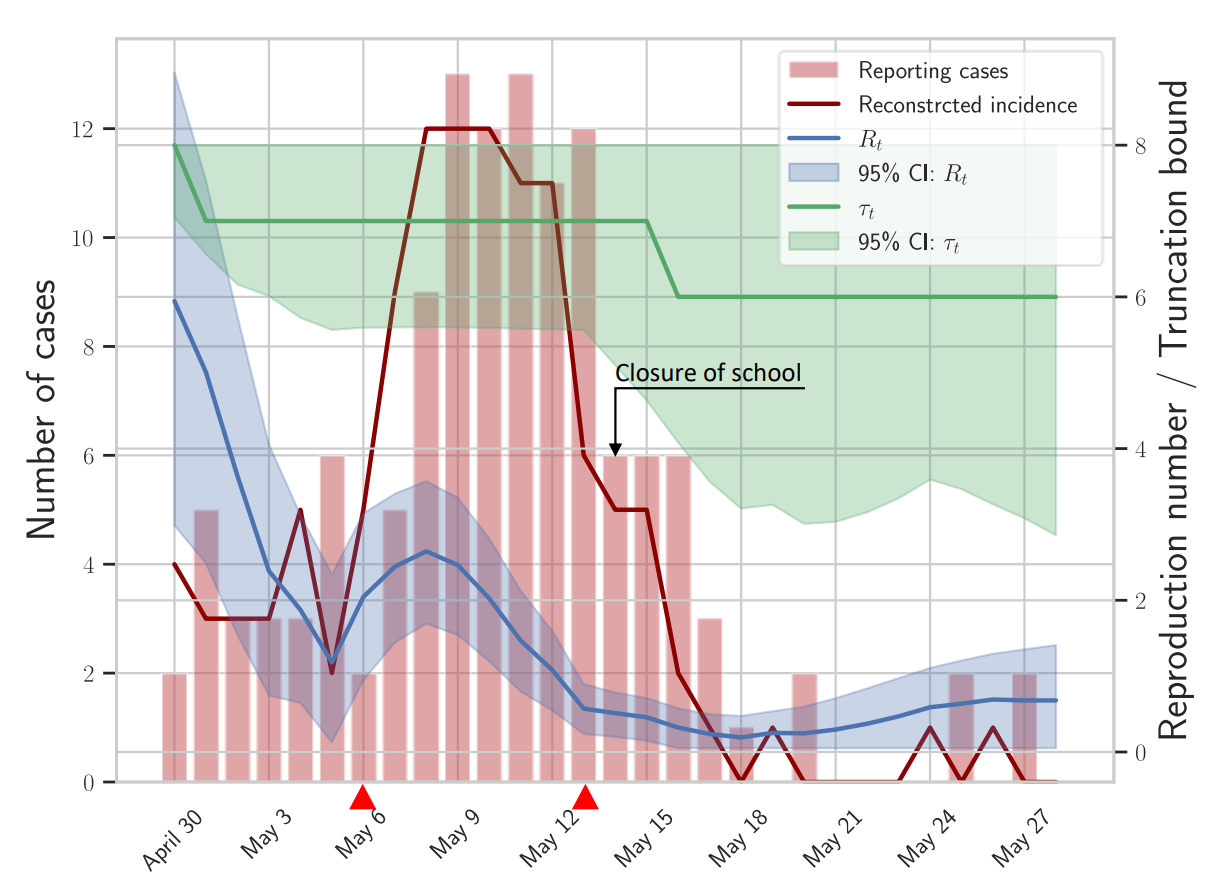
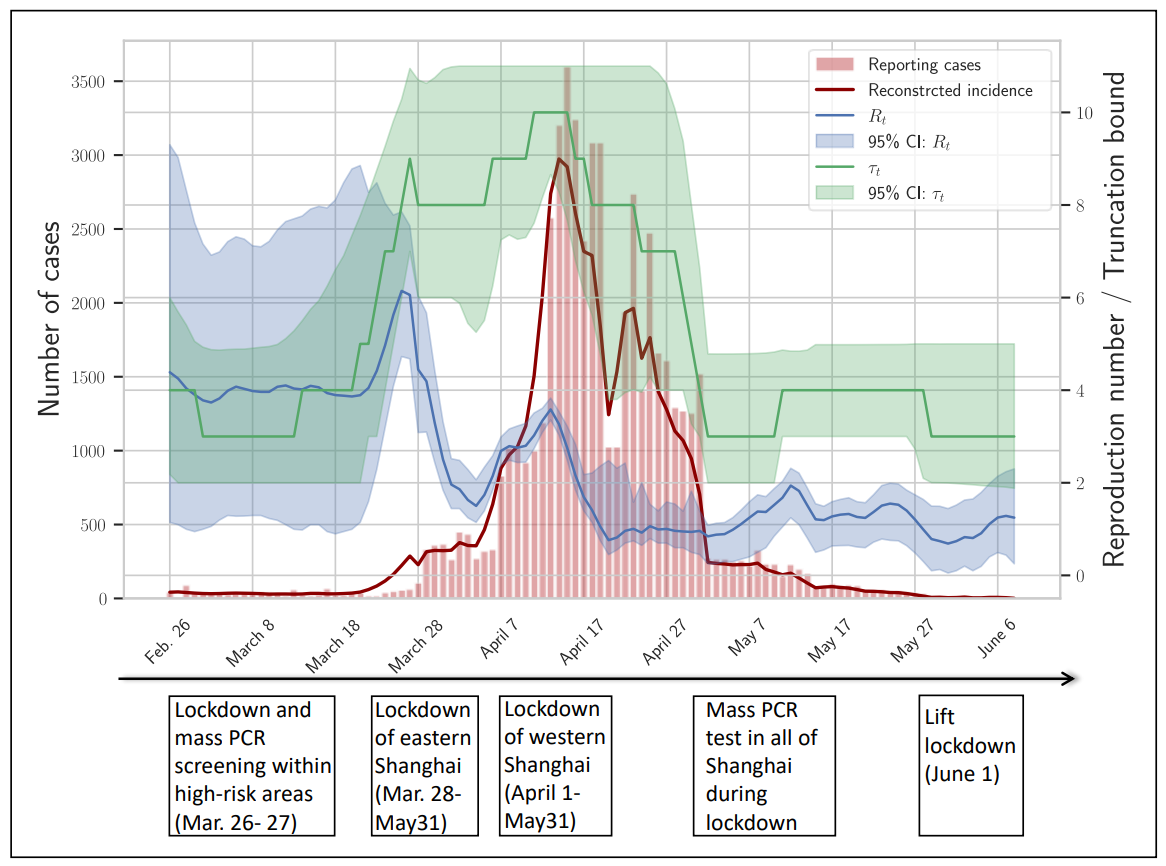
We demonstrated how to explore possible structural break points during an emerging outbreak and how epidemiological quantities could be inferred using data from outbreaks of H1N1 influenza and SARS-CoV-2 Omicron variant.
The sequence of cases, which is defined as onset of acute respiratory illness, is provided in the EpiEstim, an R software package. This data is also available as supplement material at Improved inference of time-varying reproduction numbers during infectious disease outbreaks.
Aggregated data of daily confirmed cases were gathered from John Hopkins University database.
Details of both examples are available at JupyterNotebook file in this repository.