This is the method used to generate synthetic X-ray images from CT volumes on LIDC-IDRI dataset, as introduced in the CVPR 2019 paper: X2CT-GAN: Reconstructing CT from Biplanar X-Rays with Generative Adversarial Networks.
glob, scipy, numpy, cv2, pfm, matplotlib, pydicom, SiimpleITK, re
-
Install the DRR software from here on a Windows computer, and we use 1.7.3.12-win64 version in our original work.
-
Download LIDC-IDRI dataset. (Note: Original dataset has a size of 125 GB. You could only download part of it, since this code could process it anyway)
-
In mhd_gen.py, set
-
RootPathDicom: path to LIDC-IDRI dataset
-
SaveRawData: path to save raw mhd file for ct processing in the next step
-
SaveMaskData: path to save mask that segment the lung for other part of the image by setting a threshold to HU value. Here we get mask by using
cv.threshold(slice, 0, 255, cv.THRESH_BINARY+cv.THRESH_OTSU)from opencv-pythonThen execute this file to generate mask and raw mhd file
-
-
In ctpro.py, first set data path;
- root_path : original CT volume root
- mask_root_path : CT mask without table
- save_root_path : save dir
- then execute ctpro.py to generate the normalized CT volumes.
-
In xraypro.py, again set the data path;
- root_path : the normalized data path, same as the save_root_path in ctpro.py file
- save_root_path :the Xray output path
- plasti_path : DDR software executable file location
- then execute xraypro.py to generate the synthetic Xrays.
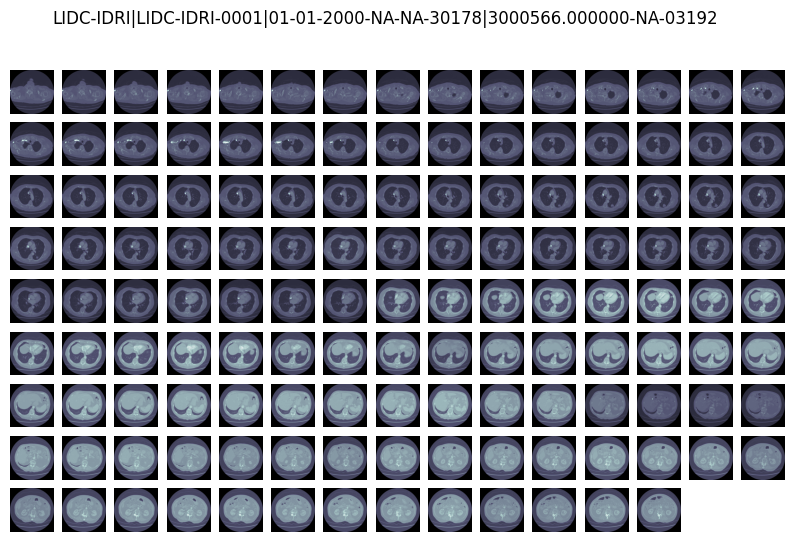
Slices of LIDC-IDRI-0001|01-01-2000-NA-NA-30178|3000566.000000-NA-03192
Generated mask by running mhd_gen.py
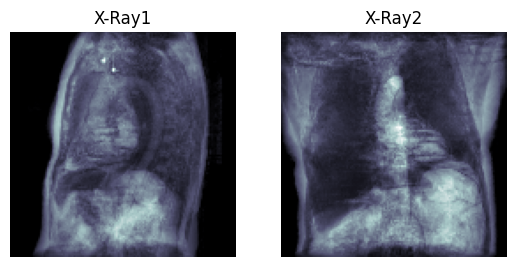
Generated X-Ray by running ctpro.py and xraypro.py