searching the ChEMBL database for molecules matching SMARTS patterns
Currently, the dependency paths in Cargo.toml are hard-coded relative paths to
other projects on my computer. You can of course edit these manually, but if you
want to use this project, let me know, and I will change them to GitHub paths.
In addition to these dependencies, you also need to have an active conda
environment with the openff-toolkit installed and a local build of
rdkit. See the Makefile for how to set your
LD_LIBRARY_PATH correctly.
cargo run --release -- \
--molecule-file chembl_33.sdf \
--search-params want.params \
--write-outputThis command reads chembl_33.sdf located in the current directory, searching
for matches to SMIRNOFF parameters listed in want.params, and writes one
pid.smiles output file for each parameter in want.params. You can obtain a
gzipped version of the ChEMBL 33 database here. However, nothing
should be ChEMBL specific, and the program should run on any valid SDF file. The
format of want.params is quite simple, one parameter identifier per line:
t18a
t18b
t87a
t123
t123a
t138a
The lines are trimmed of leading and trailing whitespace but otherwise
unaltered. The default OpenFF force field from which these parameters and their
SMIRKS are extracted is openff-2.1.0.offxml (Sage 2.1.0). You can change this
to any built-in force field or file readable by the openff-toolkit with
the --forcefield/-f flag. Similarly, if you want to look at parameters other
than ProperTorsions, you can pass a different parameter handler type via the
--parameter-type/-p flag.
By default, the search uses all available CPU cores, but you can adjust this
with the --threads/-t flag. You'll want it to be pretty high because it takes
~8 minutes on my 16 core desktop. Also be sure to use the --release flag or it
will take more like 2 hours.
Once you have output files from the main search, you can use the fingerprint
binary to process the raw SMILES output into an HTML report. Example use looks
like:
cargo run --bin fingerprint --release -- \
t18a.smiles --min-pts 10 --parameter t18aThis reads the file t18a.smiles in the current directory, generates Morgan
fingerprints with the default radius of 4 and length 1024 for each of the
corresponding molecules, computes the Tanimoto distance matrix for
the molecules, and performs a DBSCAN clustering before producing an
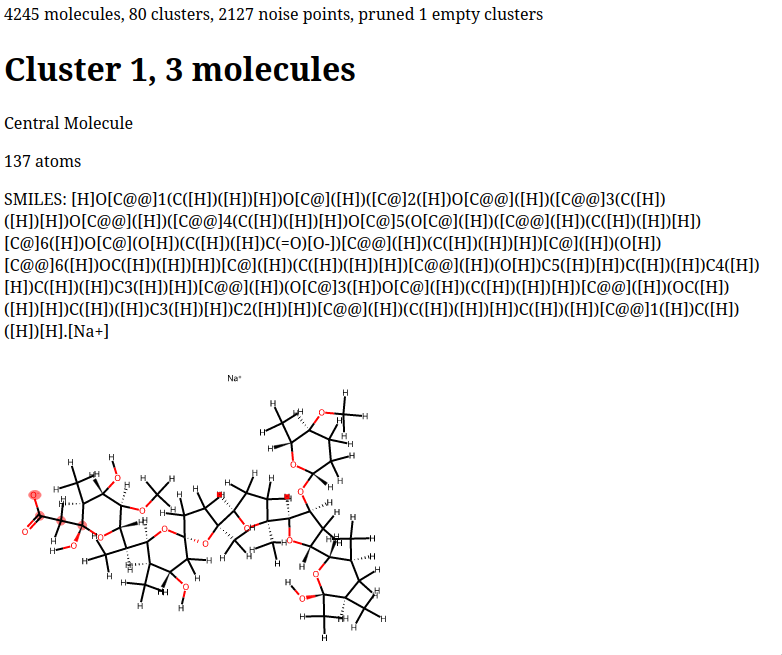
HTML report summarizing the output. You can see the top of an example output
file (for a horrifying SMILES string) below.
Again, you can modify the parameters for the clustering and drawing using
command line flags. These flags alter the behavior of the atom highlighting code
and only apply when the --parameter/-p is supplied. Otherwise, no atoms are
highlighted in the generated images.
--forcefieldchanges the OpenFF force field--parameterchanges the parameter identifier--parameter-typechanges the parameter handler type
These flags alter the parameters for the DBSCAN algorithm:
--epsilonsets the maximum cluster radius--min-ptssets the minimum cluster size
And this flag alters the Morgan fingerprinting:
--radiussets the Morgan fingerprinting radius