Mathis Petrovich · Or Litany · Umar Iqbal · Michael J. Black · Gül Varol · Xue Bin Peng · Davis Rempe
This repository hosts the official PyTorch implementation of the paper "Multi-Track Timeline Control for Text-Driven 3D Human Motion Generation" (CVPRW 2024). For more details, please visit our project webpage.
If you use our code in your research, kindly cite our work:
@inproceedings{petrovich24stmc,
title = {Multi-Track Timeline Control for Text-Driven 3D Human Motion Generation},
author = {Petrovich, Mathis and Litany, Or and Iqbal, Umar and Black, Michael J. and Varol, G{\"u}l and Peng, Xue Bin and Rempe, Davis},
booktitle = {CVPR Workshop on Human Motion Generation},
year = {2024}
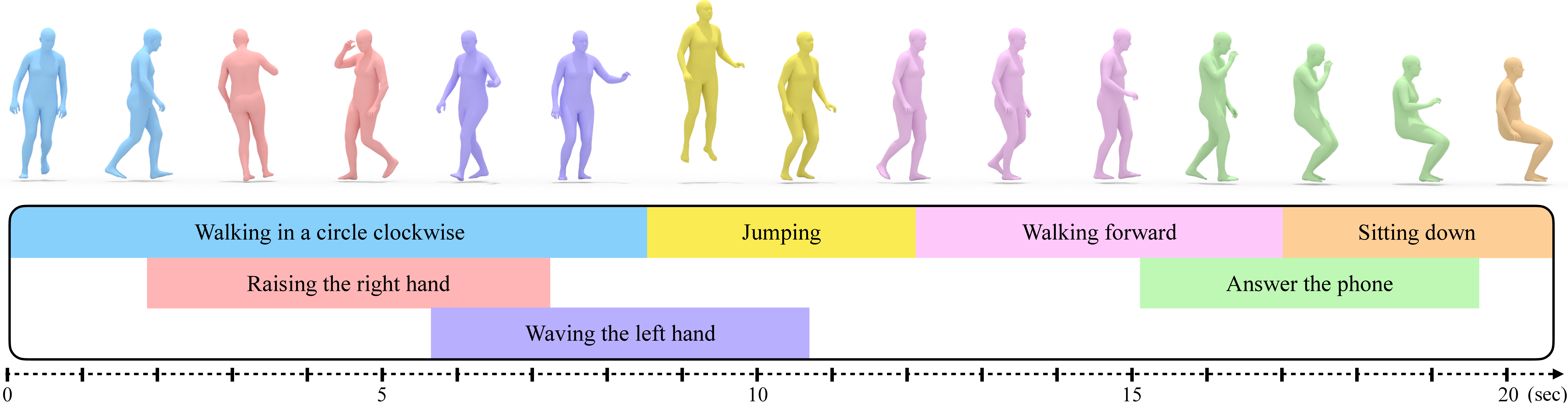
}A timeline is represented as a list of text intervals, one for each lines, as follow:
walking in a circle # 0.0 # 6.0 # legs
raising both hands in the air # 2.0 # 5.0 # right arm # left arm
Each text interval contains several pieces of information separated by #:
- text description
- start time (in seconds)
- end time (in seconds)
- list of body parts
In this code, we assume that we know the body parts, but we could also use GPT-3 (as in SINC) or learn a model to label them automatically.
Key components of the STMC algorithm are located in src/stmc.py, with additional functions in src/bptools.py. The algorithm includes:
read_timelines: Reads and parses timelines from a file.process_timeline: Preprocesses timelines as described in the paper.combine_features_intervals: Spatially and temporally combines motions (at each denoising step).
STMC is designed for integration with existing diffusion models.
In addition to the STMC algorithm, we also provide a fast diffusion model that directly generates the SMPL pose parameters.
Clone and set up the environment as follows:
git clone https://github.com/nv-tlabs/stmc
cd stmc/This code was tested with python 3.10.12, cuda 12.1 and pytorch "2.0.1+cu118".
Creation of the environnement:
# create a virtual environnement (also works with conda)
python -m venv ~/.venv/STMC
# activate the virtual environnement
source ~/.venv/STMC/bin/activate
# upgrade pip
python -m pip install --upgrade pip
# Install pytorch (see https://pytorch.org/get-started/locally)
python -m pip install torch torchvision --index-url https://download.pytorch.org/whl/cu118
# Install missing packages
python -m pip install -r requirements.txtIf it does not work well on Ubuntu, you can try installing this package:
sudo apt-get install python3.10-venvFollow the instructions below to set up the datasets within the stmc directory.
mkdir datasets
Please follow the README from TEMOS to obtain the deps folder with SMPL+H downloaded, and place deps in the stmc directory.
Click to expand
The motions all come from the AMASS dataset. Please download all "SMPL-H G" motions from the AMASS website and place them in the folder datasets/motions/AMASS.
It should look like this:
datasets/motions/
└── AMASS
├── ACCAD
├── BioMotionLab_NTroje
├── BMLhandball
├── BMLmovi
├── CMU
├── DanceDB
├── DFaust_67
├── EKUT
├── Eyes_Japan_Dataset
├── HumanEva
├── KIT
├── MPI_HDM05
├── MPI_Limits
├── MPI_mosh
├── SFU
├── SSM_synced
├── TCD_handMocap
├── TotalCapture
└── Transitions_mocapEach file contains a "poses" field with 156 (52x3) parameters (1x3 for global orientation, 21x3 for the whole body, 15x3 for the right hand and 15x3 for the left hand).
Then, launch these commands:
python prepare/amasstools/fix_fps.py
python prepare/amasstools/smpl_mirroring.py
python prepare/amasstools/extract_joints.py
python prepare/amasstools/get_smplrifke.pyClick here for more information on these commands
The script will interpolate the SMPL pose parameters and translation to obtain a constant FPS (=20.0). It will also remove the hand pose parameters, as they are not captured for most AMASS sequences. The SMPL pose parameters now have 66 (22x3) parameters (1x3 for global orientation and 21x3 for full body). It will create and save all the files in the folder datasets/motions/AMASS_20.0_fps_nh.
This command will mirror SMPL pose parameters and translations, to enable data augmentation with SMPL (as done by the authors of HumanML3D with joint positions).
The mirrored motions will be saved in datasets/motions/AMASS_20.0_fps_nh/M and will have a structure similar than the enclosing folder.
The script extracts the joint positions from the SMPL pose parameters with the SMPL layer (24x3=72 parameters). It will save the joints in .npy format in this folder: datasets/motions/AMASS_20.0_fps_nh_smpljoints_neutral_nobetas.
This command will use the joints + SMPL pose parameters (in 6D format) to create a unified representation (205 features). Please see prepare/amasstools/smplrifke_feats.py for more details.
The dataset folder should look like this:
datasets/motions
├── AMASS
├── AMASS_20.0_fps_nh
├── AMASS_20.0_fps_nh_smpljoints_neutral_nobetas
└── AMASS_20.0_fps_nh_smplrifkeClick to expand
In the stmc directory, clone the TMR repo: git clone https://github.com/Mathux/TMR.git.
It will also be useful for evaluation purposes.
Run this command to copy the processed HumanML3D text annotations. This allows us to use the HumanML3D annotations with the original AMASS sequences, as explained in the AMASS-Annotation-Unifier repo.
cp -r TMR/datasets/annotations datasets/Next, run the following command to pre-compute the CLIP embeddings (ViT-B/32):
python -m prepare.embeddingsThe folder should look like this:
datasets/annotations/humanml3d
├── annotations.json
├── splits
│ ├── all.txt
│ ├── test_tiny.txt
│ ├── test.txt
│ ├── train_tiny.txt
│ ├── train.txt
│ ├── val_tiny.txt
│ └── val.txt
└── text_embeddings
└── ViT-B
├── 32_index.json
├── 32.npy
└── 32_slice.npy
Start the training process with:
python train.pyIt will launch a training on humanml3d with the clip embeddings and with the smplrifke representation. It will save the outputs to the folder outputs/mdm-smpl_clip_smplrifke_humanml3d which we will refer to RUN_DIR.
Please run this command to download the pre-trained model:
bash prepare/download_pretrain_models.shThe model will be downloaded and saved in pretrained_models/mdm-smpl_clip_smplrifke_humanml3d.
If the download does not start ("Access denied" with gdown)
You can either:
- Click on this link and download from your web browser
- Follow this stackoverflow answer, by creating a Google token and run the command:
curl -H "Authorization: Bearer YOUR_ACCESS_TOKEN" "https://www.googleapis.com/drive/v3/files/1CNXnqxGcVHDYzjrfZOlBge7A54OfCt2d?alt=media" -o pretrained_models.tgzThen, you can extract the pretrain model by using this command:
bash prepare/extract_pretrain_models.shGenerate a 3D human motion from a timeline using the following command:
python generate.py run_dir=RUN_DIR timeline=PATHwith PATH the path to the timeline file (for example timelines/walking_circle_raising_arms.txt).
You can use device=cpu if you don't have a gpu.
Launch a demo to generate motions from timelines with the following command:
python app.pyThe parameters can be changed in app_config.py. It takes the pretrained model by default.
The MTT dataset can be found in its entirety in the file mtt/MTT.txt, or file by file in this folder mtt/MTT/.
Click to expand
It is based on the list of text prompts in the file mtt/texts.txt (which corresponds to this list of motions in AMASS mtt/motions.txt). Sampling is done by this mtt/sampling_timelines.py file (and mtt/timelines_to_mtt.py) but you don't need to run them again.
You can use this script to generate joints for all the timelines in the MTT dataset. It is required for the evaluation below.
python generate_mtt.py run_dir=RUN_DIR [value_from=XX]The parameter value_from is optional, it can be set to:
joints(by default): to extract the joints directly from the representationsmpl: to extract SMPL pose parameters from the representation and use the SMPL layer to obtain the joints
STMC can also be used in a pre-existing diffusion model, provided that it operates on the "motion space" (no latent diffusion).
Click to expand
Please follow the MDM installation instructions in their repo, and go to the repo folder:
cd /path/to/motion-diffusion-modelThis code has been tested on commit 8139dda55d90a58aa5a257ebf159b2ecfb78c632. To check the current commit, you can run git rev-parse HEAD (or run git log). Copy the additional files into the MDM code:
cp -r /path/to/stmc/integration/stmc_mdm .
cp -r /path/to/stmc/mtt .Download the pre-trained humanml_trans_enc_512 model from the README.
The following commands take this model by default --model_path=./save/humanml_trans_enc_512/model000200000.pt, but it can be changed to another as long as it is trained on Humanml3D.
To generate a motion from a timeline, you can run this command:
python -m stmc_mdm.generate --input_timeline PATHThis script behaves in the same way as MDM, and creates a folder in which the results are stored (visualisation + ``.npy'').
To generate joints from all the timelines in the MTT dataset (required for evaluation), you can run this command:
python -m stmc_mdm.generate_mttIt will create a folder with all the joints stored in ``.npy'' format. Note that the joints generated have the axis of gravity in Y (and not in Z as in Ours). This difference is handled in the evaluation script.
Click to expand
Please follow the MotionDiffuse installation instructions in their repo, and go to the "text2motion" folder in their repo folder:
cd /path/to/MotionDiffuse/text2motionThis code has been tested on commit aba848edd22133919ca96b67f4908399c67685b1. To check the current commit, you can run git rev-parse HEAD (or run git log). Copy the additional files into the MotionDiffuse code:
cp -r /path/to/stmc/integration/stmc_motiondiffuse .
cp -r /path/to/stmc/mtt .Download the pre-trained t2m_motiondiffuse model from the README.
The following commands take this model as the default --opt_path checkpoints/t2m/t2m_motiondiffuse/opt.txt, but it can be changed to another provided it is trained on Humanml3D.
To generate a motion from a timeline (stored in the ``PATH'' folder), you can run this command:
PYTHONPATH="$(dirname $0)/..":$PYTHONPATH python -u stmc_motiondiffuse/generate.py --input_timeline PATH --result_path "motion.mp4" --npy_path "joints.npy"This script behaves in a similar way to MotionDiffuse, storing the visualisation + joints in ``.npy''.
To generate joints from all the timelines in the MTT dataset (required for evaluation), you can run this command:
PYTHONPATH="$(dirname $0)/..":$PYTHONPATH python -u stmc_motiondiffuse/generate_mtt.pyIt will create a folder with all the joints stored in ``.npy'' format. Note that the joints generated have the axis of gravity in Y (and not in Z as in Ours). This difference is handled in the evaluation script.
Click to expand
We present in the paper 3 (+1) baselines.
One Text: collapse the multi-track timeline into a single text (A then B while C)Single track: collapse the multi-track timeline into a single track (with while) (corresponds to theDiffCollagemethod)SINC: only combine the motions at the end of the generationSINC lerp: same as SINC with linear interpolation to smooth the transitions
For the baselines One Text and Single track, it is implemented by pre-processing the timelines (see the mtt/create_baselines.py script).
The timelines are already pre-processed in this repo in the folder: mtt/baselines. Then, it is possible to use the STMC algorithm directly on these timelines.
For the SINC baselines, we need to do the standard diffusion process for the generation, and merge the motions by body parts at the end (and do linear interpolation if needed). There is a special method called sinc_baseline in the diffusion object to handle this.
For SINC baselines, we need to do the standard diffusion process for generation, and merge the motions by body parts at the end (+ linear interpolation for lerp). There is a special method called sinc_baseline to handle this in this model.
To generate the motions with the baseline methods on the MTT dataset, you can add a parameter to the generate_mtt script:
python generate_mtt.py run_dir=RUN_DIR baseline=XXwith XX which corresponds to:
noneby default (for STMC)onetextsingletracksincsinc_lerp
For MDM and MotionDiffuse, you should add --stmc_baseline=XX.
You should already have TMR installed in this repo (used for dataset installation). Follow their README and configure the TMR code (download the pre-trained models). Then copy these folders and files:
cd /path/to/stmc
cp -r mtt TMR/
cp mtt/motion_representation_local.py TMR/src/guofeats/The mtt/experiments.py file contains all the experiments in Table 1 of the paper. Feel free to assign the experiments variable to the list of only the generations you wish to evaluate. Then run this command to obtain the metrics (in LaTeX format):
cd TMR/
python -m mtt.eval_mttThis code, the MTT dataset, and pre-trained models are distributed under this non-commercial license. Models are trained using AMASS and HumanML3D data, which have their own respective licenses that must be followed:
Parts of this codebase are based on other repos including: