The main goals of this shootout were
- learn how to use Cython
- figure out how to optimize a Cython implementation given a well known task
The task was implementing numerical solvers for ordinary differential equations
(ODEs), namely the forward-Euler, 2nd order Runge-Kutta, and finally the 4th
order Runge-Kutta method. Results should be stored in numpy.ndarray
objects, as the implementations may be integrated into an existing toolkit to
simulate large-scale neural networks that already makes heavy use of
numpy.
The shootout does not contain an optimized variant for 1D. If the system that
you wish to integrate is only 1D, simply browse the web and take one of the
existing Cython solutions. Or quickly port the pure python version to proper
Cython. This means to add cdef everywhere where it is possible. The
speedup is well beyond 50x the original pure python speed.
Things get more interesting as soon as you have to solve an n-dimensional
system. Directly using numpy.arrays in Cython yields only mediocre
speedups, if any at all. Hence this shootout.
- pure python
- numpy ndarray
- symlink to the pure python method
- how good is Cython in translating pure python code that uses numpy
- get rid of python objects as far as possible
- store result in one huge matrix, pass around slices into that matrix
- move the problem into a class to avoid creation of temporaries
- how much speedup by reducing slice-usage where possible
- can we gain speedup by using pointers all the time it is actually possible?
$ ./shootout.py
building cython modules if necessary
running build_ext
running method 'pure python'
running method 'pure cythonized'
running method 'sliced cython'
running method 'no-slice cython'
running method 'pointer cython'
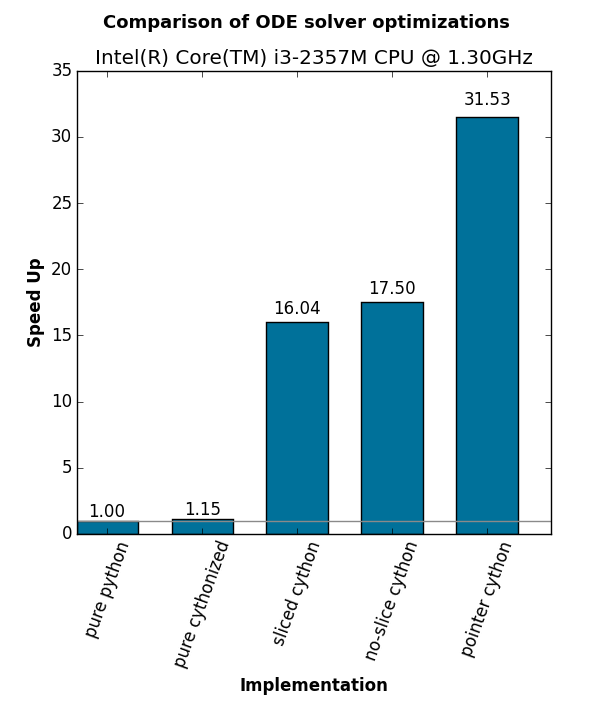
platform: Intel(R) Core(TM) i3-2357M CPU @ 1.30GHz
method runtime speed
----------------------------------------
pure python 4.6412 1.0000
pure cythonized 4.0361 1.1499
sliced cython 0.2894 16.0394
no-slice cython 0.2652 17.5029
pointer cython 0.1472 31.5325- always measure all the changes that you introduce!
- if possible, write the code that you need to speed up in C, C++, or Fortran right from the beginning and call it using Cython
- optimized Cython looks almost like C anyway...