A set of tools to impute high throughput genotyping data. Data in plink binary format is expected as input. imputation-tools will:
- Split the data by chromosomes
- Align the data according to reference strand using bcftools fixref
- Phase the data with Eagle2 and impute the data using Minimac3 using 1000 genomes reference panel
- Perform standard association test with plink2 --glm
- Visualize the GWAS results using qqman
imputation-tools was tested on Ubuntu 18.04.4 LTS. It requires a number of packages that can be installed using conda (Miniconda 3 should be sufficient).
Clone repoitory, unpack files, install environment and required packages, activate environment:
git clone https://github.com/oborisov/imputation-tools.git
cd imputation-tools
gunzip data/*gz app/*gz
conda env create --file environment.yml
conda activate imputation-tools
snakemake --config chromosome=22 bfile=data/sim1_GSA BCFTOOLS_PLUGINS=$(which bcftools | sed 's/bin\/bcftools/libexec\/bcftools/')
The running time for chromosome 22 of the included test data is approximately 90 minutes (2.50GHz CPU, 1 thread).
The following options can be passed to the imputation-tools via --config command:
- chromosome
- path to binary plink file prefix
- path to BCFTOOLS_PLUGINS, should be determined automatically based on the bcftools installation via conda
While running, imputation-tools downloads required files for QC and imputation:
- fasta file for the analyzed chromosome to align the data according to reference strand
- reference vcf file to phase and impute the analyzed chromosome
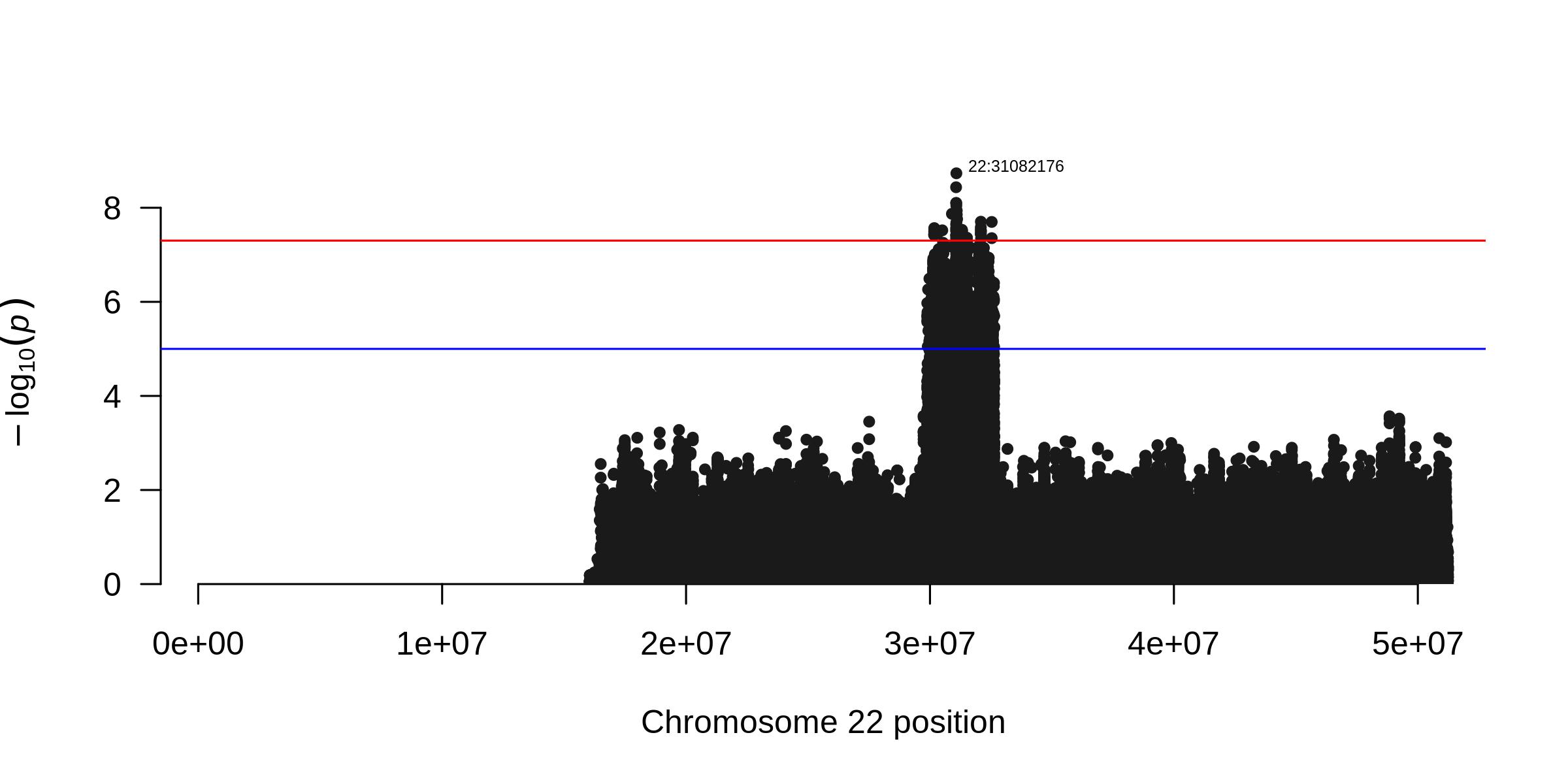
imputation-tools outputs Manhattan plot for the analyzed chromosome:
snakemake: https://snakemake.readthedocs.io/en/stable/
plink2: https://www.cog-genomics.org/plink/2.0/
bcftools: http://samtools.github.io/bcftools/bcftools.html
samtools: http://samtools.github.io/
tabix: http://www.htslib.org/doc/tabix.html
Eagle2: https://data.broadinstitute.org/alkesgroup/Eagle/
Minimac3: https://genome.sph.umich.edu/wiki/Minimac3
pull latest version of container: docker pull olegborisov/imputation-tools:latest
run application: docker run olegborisov/imputation-tools conda run -n snakemake snakemake