[ Paper]
By Min Gao, Yukun Guo, Tristan T.Hormel, Jie Wang, Elizabeth White, Dong-Wouk Park, Thomas S. Hwang, Steven T. Bailey, Yali Jia
This repo is the official implementation of "Nonperfused Retinal Capillaries - A New Method Developed on OCT and OCTA".
This software is copyrighted and may only be used for academic research.
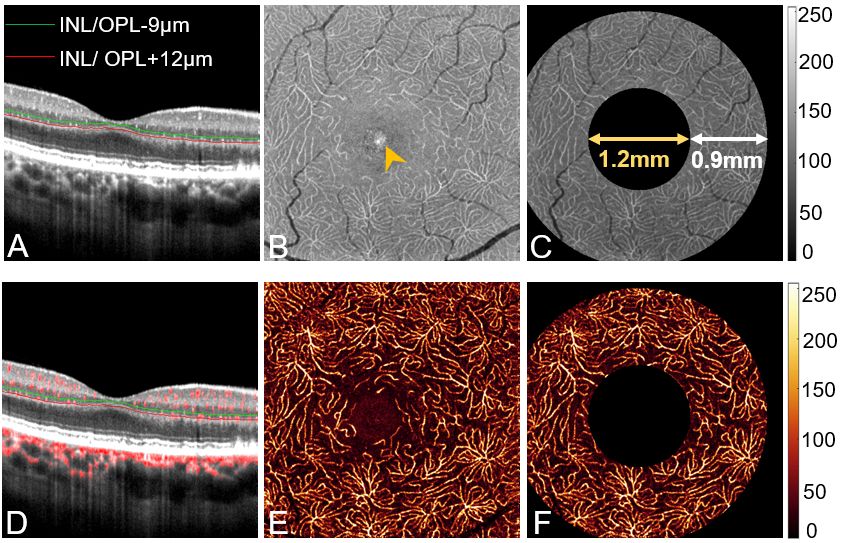
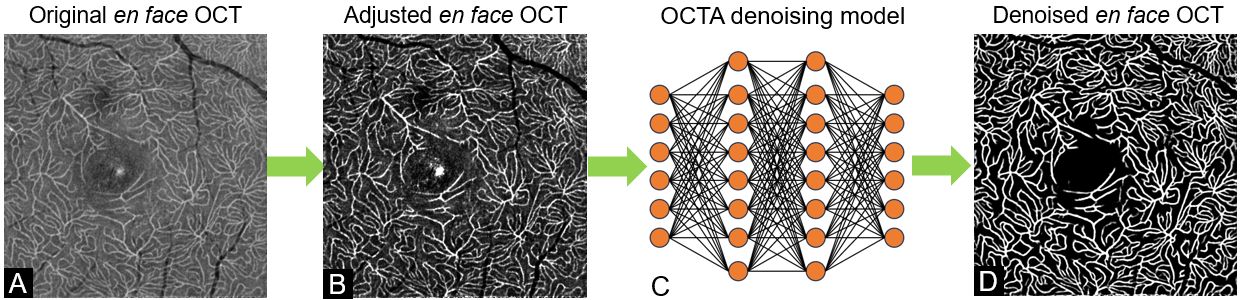
Combining co-registered optical coherence tomography (OCT) and OCT angiography (OCTA) signals allows in vivo quantification of occluded capillaries. This study aims to develop a new method to quantify nonperfused retinal capillaries (NPCs) and evaluate NPCs in eyes with age-related macular degeneration (AMD) and diabetic retinopathy (DR). We averaged multiple registered OCT/OCTA scans to create high-definition volumes. The deep capillary plexus slab was defined and segmented. A developed deep learning denoising algorithm removed tissue background noise from capillaries in en face OCT/OCTA. The algorithm segmented NPCs by identifying capillaries from OCT without corresponding flow signals in OCTA. We then investigated the relationships between NPCs and known features in AMD and DR. The segmented NPC achieved an accuracy of 88.2% compared to manual grading of DR. Compared to healthy controls, both the mean number and total length (mm) of NPCs was significantly increased in AMD and DR eyes (P < 0.001, P <0.001). Compared to early and intermediate AMD, the number and total length of NPCs were significantly higher in advanced AMD (number: P<0.001, P<0.001; total length: P = 0.002, P =0.003). Geography atrophy, macular neovascularization, drusen volume, and extrafoveal avascular area (EAA) significantly correlated with increased NPCs (P<0.05). In DR eyes, NPCs correlated with the number of microaneurysms and EAA (P<0.05). The presence of fluid did not significantly correlate with NPCs in AMD and DR. A deep learning-based algorithm can segment and quantify retinal capillaries that lack flow using colocalized OCT/OCTA. This new biomarker may be useful in AMD and DR.
git clone git@github.com:octangio/OCTA_denoising.git
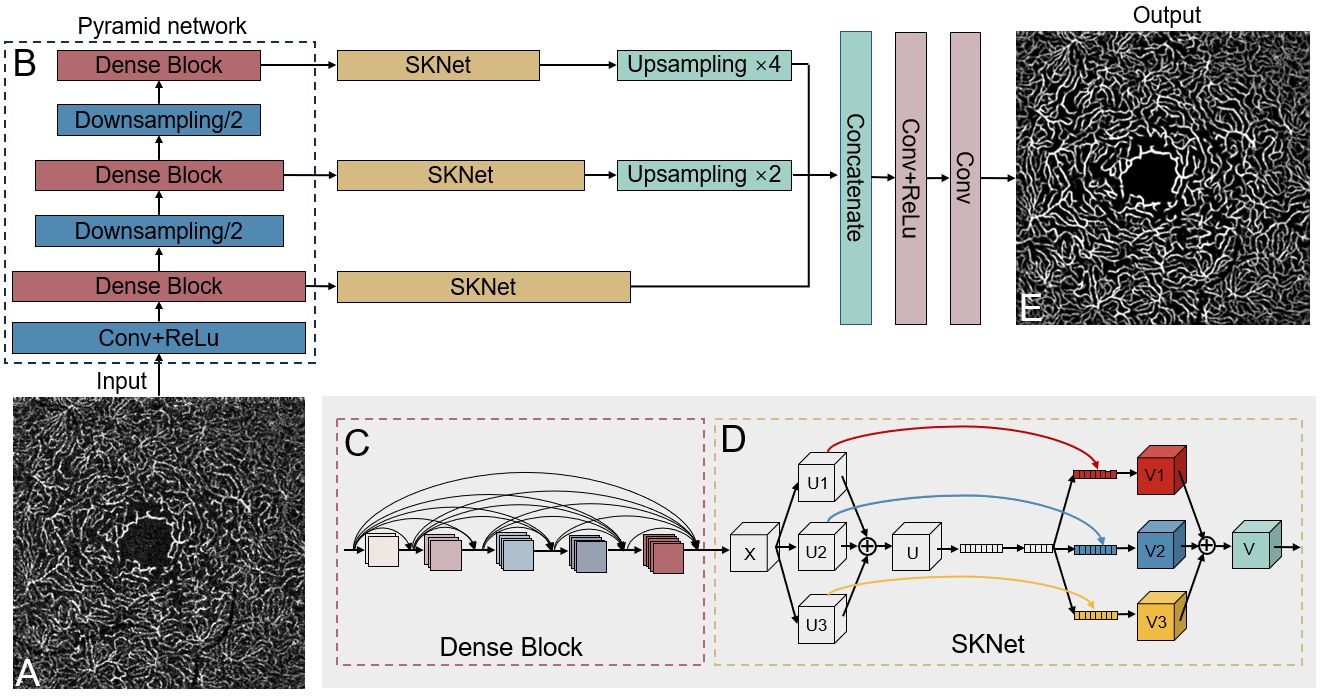
cd OCTA_denoisingpip install -r requirements.txtThe code version we used in the paper is CAVnet-1.0.
-
prepare data
The en face angiograms have normalized the range of decorrelation value (SSADA) from (0.02, 0.3) to the range of (0, 255). The data set folder should be like the following structure.
dataset | |-- train_image | | | |_list.txt | |- image_0001.png | |- image_0002.png | |- ... | |-- train_label | | | |_list.txt | |- label_0001.png | |- label_0002.png | |- ... | |-- valid_image | | | |_list.txt | |- image_0001.png | |- image_0002.png | |- ... | `-- valid_label | | |_list.txt |- label_0001.png |- label_0002.png |- ...
Then you need to generate the path list of image and label.
-
Training
python my_train.py --train_image=./dataset/train_image/_list.txt --train_label=./dataset/train_label/_list.txt --batch_size=4 --input_height=320 --input_width=320
-
Test
python predict_results.py --test_data_path=./dataset/test_data_path --save_path=./dataset/denoising_output --logdir=./logs/saved_model.hdf5