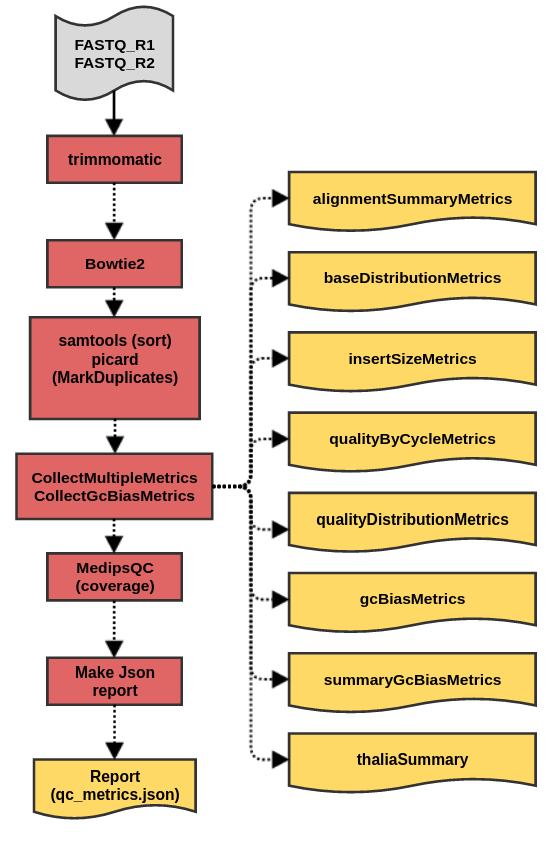
cfMedipsQC workflow produces a set of metrics files for sequencing data generated in methylation profiling of circulating Free DNA
- samtools 1.9
- bowtie2 2.1.0
- picard 2.21.2
- rstats 3.5
- python 3.7
- cfmedips 1.5.1
- trimmomatic 0.39
- bedops 2.4.37
- bc 2.1.3
java -jar cromwell.jar run cfMedipsQc.wdl --inputs inputs.json
| Parameter | Value | Description |
|---|---|---|
fastq1 |
File | Read 1 input fastq file |
fastq2 |
File | Read 2 input fastq file |
reference |
String | reference id for using with the analysis |
fastqFormat |
String | Quality encoding, default is phred33, but can be set to phred64 |
extractMedipsCounts.medips_script |
String | Path to the wrapper medips.R script |
| Parameter | Value | Default | Description |
|---|---|---|---|
window |
Int | 300 | window length, over which to assess |
| Parameter | Value | Default | Description |
|---|---|---|---|
trimming.fastq1Basename |
String | basename("~{fastq1}","_R1_001.fastq.gz") | Basename for the first fastq |
trimming.fastq2Basename |
String | basename("~{fastq2}","_R2_001.fastq.gz") | Basename for the second fastq |
trimming.headCrop |
Int | 5 | How many bases to crop |
trimming.threads |
Int | 6 | Requested CPU threads |
trimming.jobMemory |
Int | 16 | Memory (GB) allocated for this job |
trimming.timeout |
Int | 6 | Number of hours before task timeout |
trimming.modules |
String | "trimmomatic/0.39" | Module needed to run trimmomatic extract |
alignment.basename |
String | basename("~{fastq1Paired}",".R1_paired.fastq.gz") | Name to make output sam file |
alignment.threads |
Int | 8 | Requested CPU threads |
alignment.jobMemory |
Int | 16 | Memory (GB) allocated for this job |
alignment.timeout |
Int | 6 | Number of hours before task timeout |
alignment.modules |
String | "bowtie2/2.1.0 ~{referenceModule}" | Module needed to run bowtie2 alignment |
preprocessing.basename |
String | basename("~{samFile}",".sam") | Name to make output files |
preprocessing.threads |
Int | 8 | Requested CPU threads |
preprocessing.jobMemory |
Int | 16 | Memory (GB) allocated for this job |
preprocessing.timeout |
Int | 6 | Number of hours before task timeout |
preprocessing.modules |
String | "samtools/1.9 picard/2.21.2" | Module needed to run preprocessing |
alignmentMetrics.basename |
String | basename("~{dedupBam}",".sorted.dedup.bam") | Name to make output files |
alignmentMetrics.threads |
Int | 8 | Requested CPU threads |
alignmentMetrics.jobMemory |
Int | 32 | Memory (GB) allocated for this job |
alignmentMetrics.timeout |
Int | 6 | Number of hours before task timeout |
alignmentMetrics.modules |
String | "samtools/1.9 picard/2.21.2 ~{referenceModule} bc/2.1.3 rstats/3.5" | Modules needed to run alignment metrics |
splitFaiToArray.memory |
Int | 1 | Memory allocated for this job |
splitFaiToArray.timeout |
Int | 1 | Hours before task timeout |
getChromosomeLength.memory |
Int | 1 | Memory allocated for this job |
getChromosomeLength.timeout |
Int | 1 | Hours before task timeout |
getChrCoefficient.memory |
Int | 1 | Memory allocated for this job |
getChrCoefficient.timeout |
Int | 1 | Hours before task timeout |
extractMedipsCounts.basename |
String | basename("~{dedupBam}",".sorted.dedup.bam") | basename for the sample |
extractMedipsCounts.convert2bed |
String | "$BEDOPS_ROOT/convert2bed" | path to conver2bed program |
extractMedipsCounts.threads |
Int | 8 | Requested CPU threads |
extractMedipsCounts.jobMemory |
Int | 32 | Memory (GB) allocated for this job |
extractMedipsCounts.timeout |
Int | 6 | Number of hours before task timeout |
extractMedipsCounts.minCount |
Int | 5 | Minimal counts required to process a bam file |
extractMedipsCounts.modules |
String | "samtools/1.9 rstats/3.5 cfmedips/1.5.1 bedops/2.4.37" | Modules needed to run alignment metrics |
aggregateMetrics.jobMemory |
Int | 1 | Memory allocated for this job |
aggregateMetrics.timeout |
Int | 1 | Hours before task timeout |
finalMetrics.threads |
Int | 8 | Requested CPU threads |
finalMetrics.jobMemory |
Int | 16 | Memory (GB) allocated for this job |
finalMetrics.timeout |
Int | 6 | Number of hours before task timeout |
finalMetrics.modules |
String | "cfmedips/1.5.1" | Modules needed to run alignment metrics |
| Output | Type | Description | Labels |
|---|---|---|---|
outputAlignmentSummaryMetrics |
File | Metric for alignments | vidarr_label: outputAlignmentSummaryMetrics |
outputBaseDistributionMetrics |
File | Metrics for base distributions | vidarr_label: outputBaseDistributionMetrics |
outputInsertSizeMetrics |
File? | Metrics for insert size (Optional, when enough data are available) | vidarr_label: outputInsertSizeMetrics |
outputQualityByCycleMetrics |
File | Quality by cycle metrics | vidarr_label: outputQualityByCycleMetrics |
outputQualityDistributionMetrics |
File | Quality distribution metrics | vidarr_label: outputQualityDistributionMetrics |
outputGcBiasMetrics |
File | gc bias metrics | vidarr_label: outputGcBiasMetrics |
outputSummaryGcBiasMetrics |
File | Summary of gc bias metrics | |
outputThaliaSummary |
File | Summary of thalia chromosomes | vidarr_label: outputThaliaSummary |
outputDedupBam |
File | De-duplicated bam file | vidarr_label: outputDedupBam |
outputqcMetrics |
File | Final output | vidarr_label: outputqcMetrics |
This section lists commands run by cfMedipsQC workflow
- Running cfMedipsQC
cfMedipsQC is designed to produce several QC metrics using samtools and picard. Runs it's own alignment with bowtie2
grep -w ^~{chromosome} REF_FAI | cut -f 2 | awk '{print int(($1/~{LARGEST_CHR_BASES} + 0.1) * 10)'
set -euo pipefail
trimmomatic PE FASTQ_R1 FASTQ_R2 \
"-phred33 \ (can be set to phred64)
"FASTQ_R1_BASENAME.R1_paired.fastq.gz" "FASTQ_R1_BASENAME.R1_unpaired.fastq.gz" "FASTQ_R2_BASENAME.R2_paired.fastq.gz" "FASTQ_R2_BASENAME.R2_unpaired.fastq.gz" \
HEADCROP:5 (how many bases to crop, configurable)
set -euo pipefail
bowtie2 -p 8 -x REFEFENCE_FASTA \
-1 FASTQ_R1 \
-2 FASTQ_R2 \
-S "BASENAME.sam"
set -euo pipefail
samtools view -bS SAM_FILE | samtools sort -o "BASENAME.sorted.bam"
java -jar picard.jar MarkDuplicates \
I="BASENAME.sorted.bam" \
O="BASENAME.sorted.dedup.bam" \
M="BASENAME.sorted.dedup.metrics" \
ASSUME_SORTED=true \
CREATE_INDEX=true \
VALIDATION_STRINGENCY=SILENT \
REMOVE_DUPLICATES=false
set -euo pipefail
java -jar picard.jar CollectMultipleMetrics \
R=REFERENCE_GENOME\
I=DEDUPLICATED_BAM \
O=BASENAME \
VALIDATION_STRINGENCY=SILENT
java -jar picard.jar CollectGcBiasMetrics \
R=REFERENCE_GENOME \
I=DEDUPLICATED_BAM \
O="BASENAME.gc_bias_metrics.txt" \
S="BASENAME.summary_gc_bias_metrics.txt" \
CHART="BASENAME.gc_bias_metrics.pdf"
samtools view DEDUPLICATED_BAM | cut -f 3 | sort | uniq -c | sort -nr | sed -e 's/^ *//;s/ /\t/' | awk 'OFS="\t" {print $2,$1}' | sort -n -k1,1 > thalia.counts
total=$(samtools view DEDUPLICATED_BAM | wc -l)
if [[ $(cat thalia.counts | grep "^\*" | cut -f2) == "" ]]; then unmap=0; else unmap=$(cat thalia.counts | grep "^\*" | cut -f2); fi
if [[ $(cat thalia.counts | grep F19K16 | cut -f2) == "" ]]; then methyl=0; else methyl=$(cat thalia.counts | grep F19K16 | cut -f2); fi
if [[ $(cat thalia.counts | grep F24B22 | cut -f2) == "" ]]; then unmeth=0; else unmeth=$(cat thalia.counts | grep F24B22 | cut -f2); fi
if [[ $total == "" ]]; then pct_thalia=0; else pct_thalia=$(echo "scale=3; ($methyl + $unmeth)/$total * 100" | bc -l); fi
if [[ -z $pct_thalia ]]; then pct_thalia="0"; fi
if [[ $methyl == 0 && $unmeth == 0 ]]; then bet_thalia=0; else bet_thalia=$(echo "scale=3; $methyl/($methyl + $unmeth)" | bc -l); fi
if [[ -z $bet_thalia ]]; then bet_thalia="0"; fi
echo -e "total\tunmap\tmethyl\tunmeth\tPCT_THALIANA\tTHALIANA_BETA" > thalia_summary.txt
echo -e "$total\t$unmap\t$methyl\t$unmeth\t$pct_thalia\t$bet_thalia" >> thalia_summary.txt
cut -f 1 ~{refFai} | uniq | grep -v _ | grep -v M | grep ^chr
grep -w ^~{chromosome} ~{refFai} | cut -f 2
set -euo pipefail
cat ~{sep=" " coverageWindowsFiles} > "coverage_windows.tmp"
head -n 1 coverage_windows.tmp > coverage_windows.txt
grep -v ^sample coverage_windows.tmp | awk '{sum1+=$1;sum2+=$2;sum3+=$3;sum4+=$4;sum5+=$5} END{OFS="\t";print sum1,sum2,sum3,sum4,sum5;}' >> coverage_windows.txt
cat ~{sep=" " enrichmentDataFiles} > "enrichment_data.tmp"
head -n 1 enrichment_data.tmp > enrichment_data.txt
grep -v regions enrichment_data.tmp | awk '{sum1+=$2;sum2+=$3;sum3+=$4;sum4+=$5;sum5+=$6;sum6+=$7;sum7+=$8;sum8+=$9;sum9+=$10;sum10+=$11;sum11+=$12;sum12+=$13;sum13+=$14;count+=1}END{OFS="\t";print $1, $2, sum2, sum3, sum4, sum5/count, sum6/count, sum7/count, sum8/count, sum9/count, sum10/count, sum11/count, sum12/count, sum13/count}' >> enrichment_data.txt
cat ~{sep=" " coverageCountsFiles} > "coverage_counts.tmp"
head -n 1 coverage_counts.tmp > coverage_counts.txt
tail +1 coverage_counts.tmp | awk -F'\t' '{sum1+=$2;sum2+=$3} END{OFS="\t";print $1,sum1,sum2;}' >> coverage_counts.txt
cat ~{sep=" " saturationMetricsFiles} > "saturation_metrics.tmp"
head -n 1 saturation_metrics.tmp > saturation_metrics.txt
grep -v maxEstCorReads saturation_metrics.tmp | awk '{sum1+=$2;sum2+=$3;sum3+=$4;sum4+=$5;count+=1}END{OFS="\t";print $1, sum1, sum2/count, sum3, sum4/count}' >> saturation_metrics.txt
cat ~{sep=" " nameFiles} > "name.tmp"
head -n 1 name.tmp > name.txt
grep -v samples name.tmp | sort -u >> name.txt
reference=~{reference}
if [[ $reference == hg19 ]]; then
bsGenome=BSgenome.Hsapiens.UCSC.hg19
elif [[ $reference == hg38 ]]; then
bsGenome=BSgenome.Hsapiens.UCSC.hg38
else
echo "Unsupported Reference $reference"
exit
fi
set -euo pipefail
samtools view -h ~{dedupBam} ~{chromosome}:1-~{chromosomeLength} -b > "~{chromosome}.dedup.bam"
READ_COUNT=$(samtools view ~{chromosome}.dedup.bam | wc -l)
if (( $READ_COUNT < MIN_COUNT )); then
touch coverage_windows.txt
touch name.txt
touch coverage_counts.txt
touch enrichment_data.txt
touch genome_count.txt
touch MEDIPS_window_per_chr.csv
touch saturation_metrics.txt
touch medips.bed
else
Rscript MEDIPS_SCRIPT.R \
--basedir . \
--bamfile CHROM##.dedup.bam
--samplename BASENAME \
--BSgenome bsGenome \
--chromosome CHROM## \
--ws WINDOW\
--outdir .
NAME=""
count0=$(awk '$1 == 0' genome_count.txt | wc -l)
count1=$(awk '$1 >= 1' genome_count.txt | wc -l)
count10=$(awk '$1 >= 10' genome_count.txt | wc -l)
count50=$(awk '$1 >= 50' genome_count.txt | wc -l)
count100=$(awk '$1 >= 100' genome_count.txt | wc -l)
echo -e "sample\tcount0\tcount1\tcount10\tcount50\tcount100" > coverage_windows.txt
echo -e "$NAME\t$count0\t$count1\t$count10\t$count50\t$count100" >> coverage_windows.txt
echo -e "samples\n~{basename}" > name.txt
CONVERT2BED_EXECUTABLE -d --input wig < medips.wig > medips.bed
fi
set -euo pipefail
txt-to-json.py -n FILE_NAME \
-e ENRICHMENT_DATA \
-c COVERAGE_DATA \
-w COVERAGE_WINDOW \
-s SATURATION_DATA \
-d DEDUPLICATION_DATA \
-u GCBIAS_DATA \
-a ALIGNMENT_SUMMARY_DATA \
-t TALIA_SUMMARY
For support, please file an issue on the Github project or send an email to gsi@oicr.on.ca .
Generated with generate-markdown-readme (https://github.com/oicr-gsi/gsi-wdl-tools/)