The Delly workflow produces a set of vcf files with different types of structural variant calls: Translocations, Deletions, Inversions and Duplications It uses .bam files as input. The below graph describes the process:
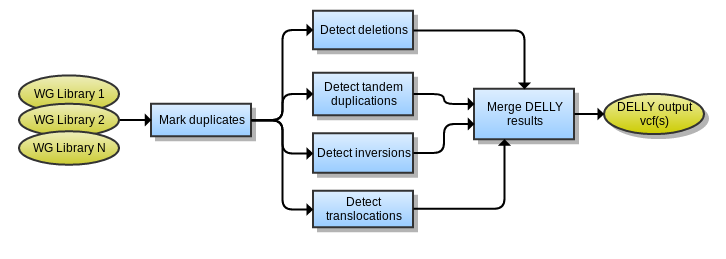
The expected inputs for the DELLY tool are aligned sequence (bam format), properly sorted and indexed, with marked duplicates.
Picard Tools MarkDuplicates is used to flag reads as PCR or optical duplicates and is activated by default. If providing bam files with duplicates marked, this can be disabled.
java -jar MarkDuplicates.jar
INPUT=sample.bam
OUTPUT=sample.dedup.bam
METRICS_FILE=sample.metrics
delly
-t DEL
-x excludeList.tsv
-o sample.jumpy.bam
-q 0
-g hn19.fa
sample.bam
delly
-t DUP
-x excludeList.tsv
-o sample.jumpy.bam
-q 0
-g hn19.fa
sample.bam
delly
-t INV
-x excludeList.tsv
-o sample.jumpy.bam
-q 0
-g hn19.fa
sample.bam
Detect translocations
delly
-t TRA
-x excludeList.tsv
-o sample.jumpy.bam
-q 0
-g hn19.fa
sample.bam
Each DELLY tool produces several files, which will all need to be merged together after the chromosomes are finished processing. The output format is described on the DELLY webpage. The merging script may require a small parser to combine the output from multiple runs in together. Merge DELLY results with vcftools
java -jar cromwell.jar run delly.wdl --inputs inputs.json
| Parameter | Value | Description |
|---|---|---|
inputTumor |
File | Tumor input .bam file. |
outputFileNamePrefix |
String | Output prefix to be used with result files. |
reference |
String | the reference genome for input sample |
| Parameter | Value | Default | Description |
|---|---|---|---|
inputNormal |
File? | None | Normal input .bam file. |
markdup |
Boolean | true | A switch between marking duplicate reads and indexing with picard. |
| Parameter | Value | Default | Description |
|---|---|---|---|
dupmarkBam.jobMemory |
Int | 20 | memory allocated for Job |
dupmarkBam.timeout |
Int | 20 | Timeout in hours |
dupmarkBam.modules |
String | "java/8 picard/2.19.2" | Names and versions of modules for picard-tools and java |
runDelly.mappingQuality |
Int | 30 | defines quality threshold for reads to use in calling SVs |
runDelly.jobMemory |
Int | 16 | memory allocated for Job |
runDelly.timeout |
Int | 20 | Timeout in hours |
mergeAndZipALL.modules |
String | "bcftools/1.9 vcftools/0.1.16 tabix/0.2.6" | Names and versions of modules for picard-tools and java |
mergeAndZipALL.variantSupport |
Int | 0 | Paired-end support for structural variants, in pairs. Default is 10 |
mergeAndZipALL.jobMemory |
Int | 10 | memory allocated for Job |
mergeAndZipFiltered.modules |
String | "bcftools/1.9 vcftools/0.1.16 tabix/0.2.6" | Names and versions of modules for picard-tools and java |
mergeAndZipFiltered.variantSupport |
Int | 0 | Paired-end support for structural variants, in pairs. Default is 10 |
mergeAndZipFiltered.jobMemory |
Int | 10 | memory allocated for Job |
| Output | Type | Description | Labels |
|---|---|---|---|
mergedIndex |
File | tabix index of the vcf file containing all structural variant calls | vidarr_label: mergedIndex |
mergedVcf |
File | vcf file containing all structural variant calls | vidarr_label: mergedVcf |
mergedFilteredIndex |
File? | tabix index of the filtered vcf file containing structural variant calls | vidarr_label: mergedFilteredIndex |
mergedFilteredVcf |
File? | filtered vcf file containing structural variant calls | vidarr_label: mergedFilteredVcf |
mergedFilteredPassIndex |
File? | tabix index of the filtered vcf file containing PASS structural variant calls | vidarr_label: mergedFilteredPassIndex |
mergedFilteredPassVcf |
File? | filtered vcf file containing PASS structural variant calls | vidarr_label: mergedFilteredPassVcf |
This section lists command(s) run by delly workflow
- Running delly
SV calling workflow
This is a job which can be optional:
java -Xmx[JOB_MEMORY-8]G -jar picard.jar MarkDuplicates
TMP_DIR=picardTmp
ASSUME_SORTED=true
VALIDATION_STRINGENCY=LENIENT
OUTPUT=INPUT_BAM_BASENAME_dupmarked.bam
INPUT=INPUT_BAM
CREATE_INDEX=true
METRICS_FILE=INPUT_BAM_BASENAME.mmm
delly call -t DELLY_MODE
-x EXCLUDE_LIST
-o SAMPLE_NAME.DELLY_MODE.CALL_TYPE.bcf
-q MAPPING_QUALITY
-g REF_FASTA
INPUT_BAM
Optional post-filtering if we need somatic variants:
echo "Somatic mode requested, will run delly filtering for somatic SVs"
bcftools view SAMPLE_NAME.DELLY_MODE.CALL_TYPE.bcf | grep ^# | tail -n 1 |
sed 's/.*FORMAT\t//' | awk -F "\t" '{print $1"\ttumor";print $2"\tcontrol"}' > samples.tsv
delly filter -f somatic -o SAMPLE_NAME.DELLY_MODE.CALL_TYPE.bcf -s samples.tsv
bcftools view SAMPLE_NAME.DELLY_MODE.CALL_TYPE_filtered.bcf |
bgzip -c > SAMPLE_NAME.DELLY_MODE.CALL_TYPE_filtered.vcf.gz
bcftools view SAMPLE_NAME.DELLY_MODE.CALL_TYPE.bcf | bgzip -c > SAMPLE_NAME.DELLY_MODE.CALL_TYPE.vcf.gz
tabix -p vcf SAMPLE_NAME.DELLY_MODE.CALL_TYPE.vcf.gz
tabix -p vcf SAMPLE_NAME.DELLY_MODE.CALL_TYPE_filtered.vcf.gz
vcf-concat INPUT_VCFS | vcf-sort | bgzip -c > SAMPLE_NAME.DELLY_MODE.CALL_TYPE_PREFIX.delly.merged.vcf.gz
tabix -p vcf SAMPLE_NAME.DELLY_MODE.CALL_TYPE_PREFIX.delly.merged.vcf.gz
bcftools view -i "%FILTER='PASS' & INFO/PE>~{variantSupport}" SAMPLE_NAME.DELLY_MODE.CALL_TYPE_PREFIX.delly.merged.vcf.gz -Oz -o SAMPLE_NAME.DELLY_MODE.CALL_TYPE_PREFIX.delly.merged.pass.vcf.gz
tabix -p vcf SAMPLE_NAME.DELLY_MODE.CALL_TYPE_PREFIX.delly.merged.pass.vcf.gz
For support, please file an issue on the Github project or send an email to gsi@oicr.on.ca .
Generated with generate-markdown-readme (https://github.com/oicr-gsi/gsi-wdl-tools/)