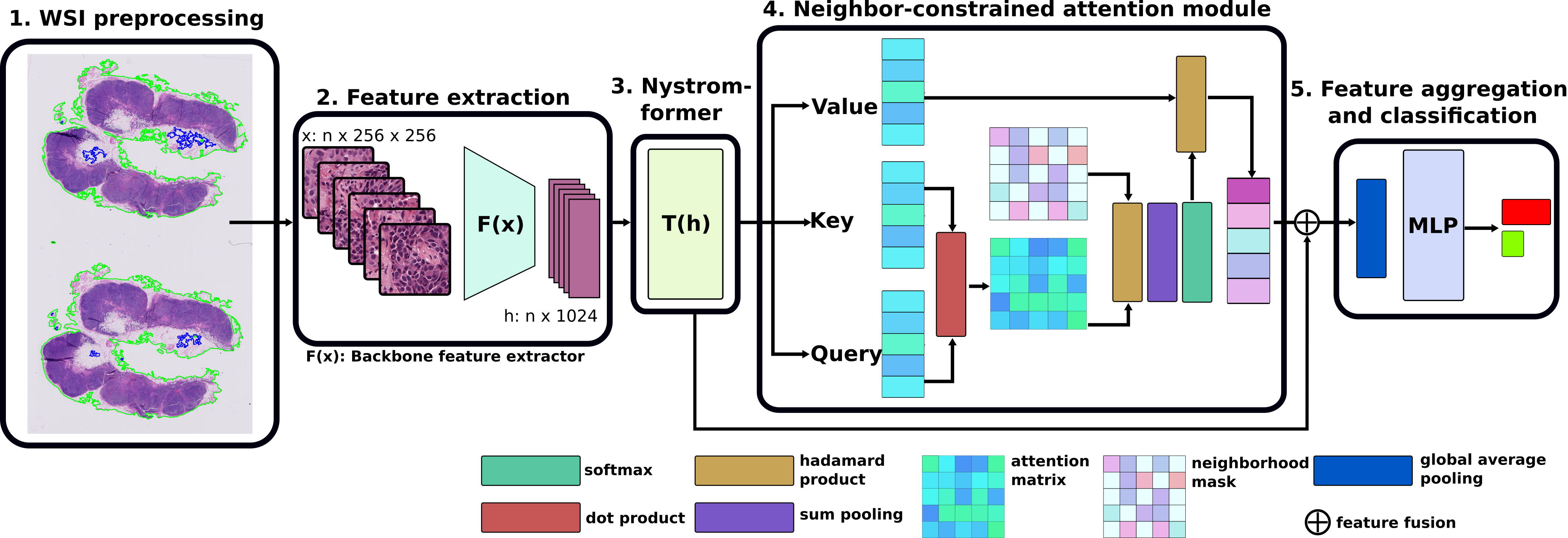
CAMIL: Context-Aware Multiple Instance Learning for Cancer Detection and Subtyping in Whole Slide Images (ICLR 2024 spotlight)
Tensorflow implementation for the multiple instance learning model described in the paper CAMIL (work in progress)
Two separate conda environments are used for different stages of the workflow:
dl_torchfor WSI preprocessing and feature extractionalmafor model training and evaluation
To create and activate these environments, run the following commands:
$ conda env create --name torch_env --file torch_env.yml
$ conda activate torch_envwe follow the CLAM's WSI processing solution (https://github.com/mahmoodlab/CLAM)
# WSI Segmentation and Patching
python create_patches_fp.py --source DATA_DIRECTORY --save_dir PATCHES_RESULTS_DIRECTORY --patch_size 256 --preset bwh_biopsy.csv --seg --patch --stitchThis script computes features using pre-trained weights and saves the results in the specified output directory
# WSI Segmentation and Patching
python feature_extractor/compute_feats.py --weights weight_dir/*.pth --dataset "PATCHES_RESULTS_DIRECTORY/*" --output FEAT_RESULTS_DIRECTORY --slide_dir DATA_DIRECTORY weight_dir: Directory containing checkpoints, one from the TCGA-NSCLC and the other from Calmeyon-16.output_dir: Directory where the H5 files are stored.slide_dir: Directory where the slides are stored.
You can download the precomputed features here: Link to Google Drive
This script computes features using pre-trained weights and saves the results in the specified output directory
python run.py --experiment_name EXP_NAME --epoch 30 --feature_path FEAT_RESULTS_DIRECTORY --label_file LABEL_FILE --csv_file SPLIT_DIR --save_dir WEIGHT_DIRexperiment_name: The name of the experiment.epoch: The number of training epochs.feature_path: The path where the features are stored.label_file: The path to the CSV file containing labels.csv_file: The path to the CSV file containing data splits.save_dir: The directory where the weights are saved stored.
The label file should be a CSV file with the following comma-separated fields: case_id, slide_id, label, slide_label
This script computes features using pre-trained weights and saves the results in the specified output directory
python run.py --experiment_name EXP_NAME --test --feature_path FEAT_RESULTS_DIRECTORY --label_file LABEL_FILE --csv_file SPLIT_DIR --save_dir WEIGHT_DIRexperiment_name: The name of the experiment.feature_path: The path where the features are stored.label_file: The path to the CSV file containing labels.csv_file: The path to the CSV file containing data splits.save_dir: The directory where the weights are saved stored.test: Flag indicating the test stage
If you use this code, please cite our work using:
@inproceedings{
fourkioti2024camil,
title={{CAMIL}: Context-Aware Multiple Instance Learning for Cancer Detection and Subtyping in Whole Slide Images},
author={Olga Fourkioti and Matt {De Vries} and Chris Bakal},
booktitle={The Twelfth International Conference on Learning Representations},
year={2024},
url={https://openreview.net/forum?id=rzBskAEmoc}
}