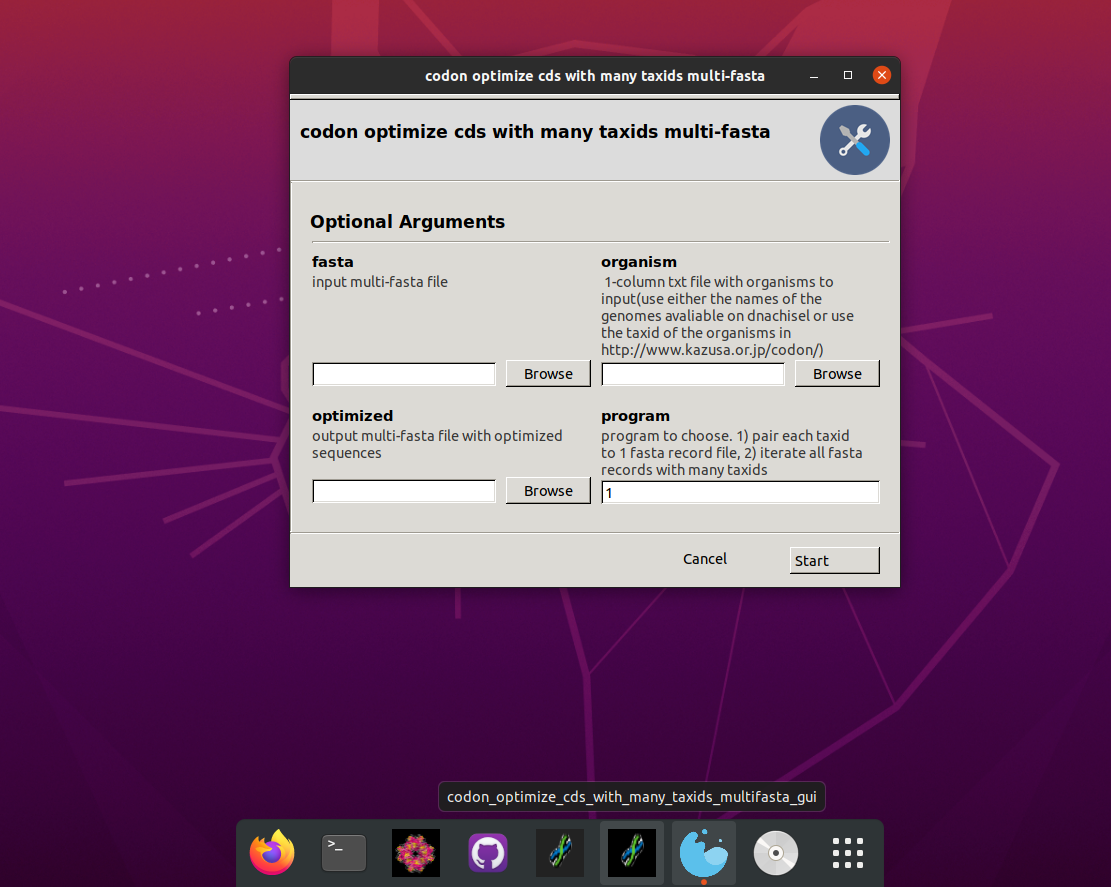
Linux GUI app to codon optimize a multi-fasta file with coding sequences with multiple taxonomy ids
- linux OS (tested on ubuntu 20.04 LTS)
- python 3.8.2 or later
- anaconda or miniconda
- Gooey
conda install -c conda-forge gooey(if you get errors during installation or runtime try installing gtksudo apt-get install libgtk-3-dev) - biopython
pip install biopython - pandas
pip install pandas - dnachisel
pip install dnachisel
- open a linux text editor and edit in the python file the shebang line (to find where python is type in the terminal
which python3) - convert to executable with
chmod +x codon_optimize_cds_with_many_taxidS_singlefasta_gui.py - open a linux text editor and edit the PATHs in the
.desktopfile - move the file to
/usr/share/applications - reboot your machine
The output multi fasta file will have fasta records with fasta headers:
- the fasta headers of the input sequences
- the name or taxonomy id of the organism to codon optimize for(as suffix)