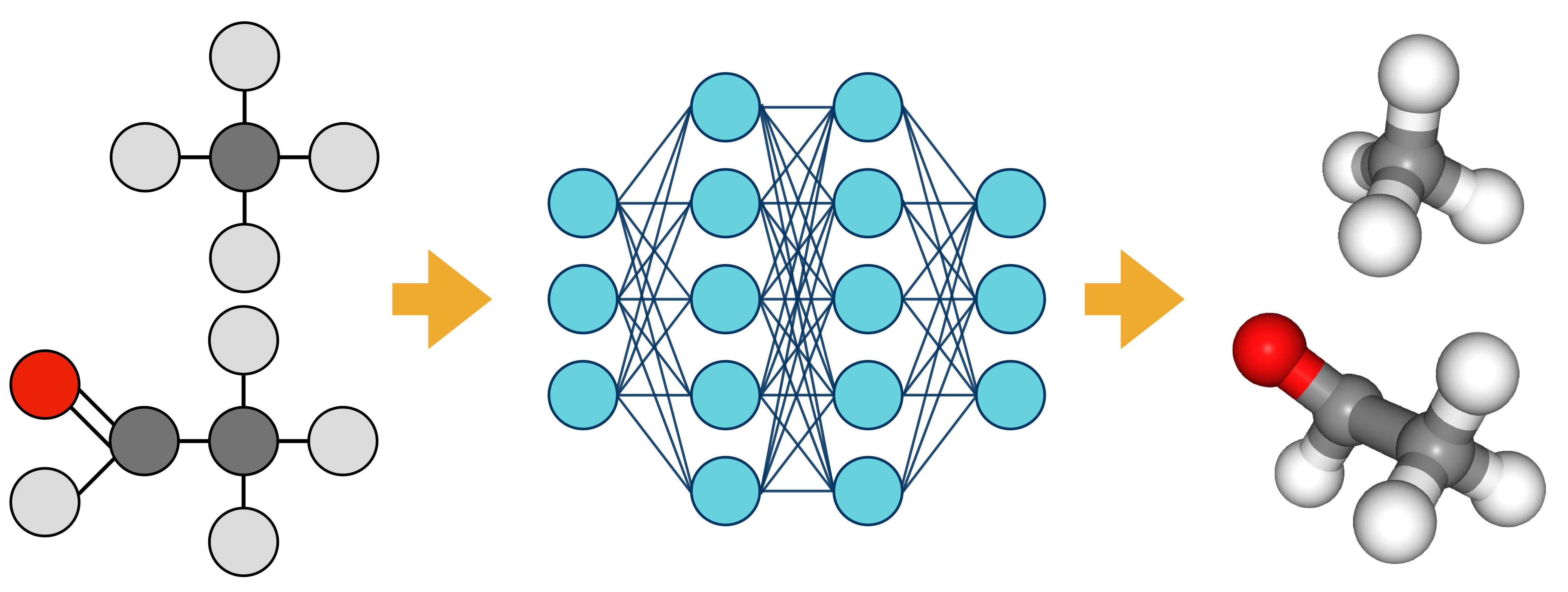
transBG is a deep-learning transferable generator of physically realistic conformers for small molecules. transBG is a Boltzmann Generator based on normalizing flows conditioned on a molecular representation. This model is auto-regressive and the position of the atoms in a molecule are generated using internal coordinates.
The following versions of transBG exist in this repository:
- v1.b is the label corresponding to the first beta version.
- Anaconda or Miniconda with Python 3.9.
- CUDA-enabled GPU.
The ./environments/transBG-env.yml file lists all the packages required for transBG to run. A virtual environment can be easily created using the YAML file and conda by typing into the terminal:
conda env create -f environments/transBG_env.yml
Then, to activate the environment:
conda activate transBG_env
The two learning strategies, likelihood- and energy-based learning, can be performed using the script submit_transBG.py. Jobs can be run using your local computer or SLURM.
The (hyper-)parameters and dataset paths/indexes can be specified at ./parameters.py.
The sdf files corresponding to the QM9 dataset molecules can be downloaded using ./preprocessing/download_qm9.py. However, the xyz-files are also needed. These latter can be downloaded from kaggle. Moreover, the conformations obtained by molecular dynamics simulations can be found at ./datasets/QM9/qm9_conformations.
@JuanViguera and @psolsson.
Contributions are welcome in the form of issues or pull requests. To report a bug, please submit an issue. Thank you to everyone who has used the code and provided feedback thus far.
If you use transBG in your research, please reference our not_yet_available.
The reference in BibTex format are available below:
transBG was initially developed in Juan Viguera Diez's Master's Thesis. His (and Sara Romeo's ) memmory can be found at
@Article {Boltzmann_gen,
author = {No{\'e}, Frank and Olsson, Simon and K{\"o}hler, Jonas and Wu, Hao},
title = {Boltzmann generators: Sampling equilibrium states of many-body systems with deep learning},
volume = {365},
number = {6457},
elocation-id = {eaaw1147},
year = {2019},
doi = {10.1126/science.aaw1147},
publisher = {American Association for the Advancement of Science},
issn = {0036-8075},
URL = {https://science.sciencemag.org/content/365/6457/eaaw1147},
eprint = {https://science.sciencemag.org/content/365/6457/eaaw1147.full.pdf},
journal = {Science}
}
transBG is licensed under the MIT license and is free and provided as-is.