This repository contains the code for Deterministic Neural Networks with Appropriate Inductive Biases Capture Epistemic and Aleatoric Uncertainty.
If the code or the paper has been useful in your research, please add a citation to our work:
@article{mukhoti2021deterministic,
title={Deterministic Neural Networks with Appropriate Inductive Biases Capture Epistemic and Aleatoric Uncertainty},
author={Mukhoti, Jishnu and Kirsch, Andreas and van Amersfoort, Joost and Torr, Philip HS and Gal, Yarin},
journal={arXiv preprint arXiv:2102.11582},
year={2021}
}
The code is based on PyTorch and requires a few further dependencies, listed in environment.yml. It should work with newer versions as well.
For OoD detection, you can train on CIFAR-10/100. You can also train on Dirty-MNIST by downloading Ambiguous-MNIST (amnist_labels.pt and amnist_samples.pt) from here and using the following training instructions.
In order to train a model for the OoD detection task, use the train.py script. Following are the main parameters for training:
--seed: seed for initialization
--dataset: dataset used for training (cifar10/cifar100/dirty_mnist)
--dataset-root: /path/to/amnist_labels.pt and amnist_samples.pt/ (if training on dirty-mnist)
--model: model to train (wide_resnet/vgg16/resnet18/resnet50/lenet)
-sn: whether to use spectral normalization (available for wide_resnet, vgg16 and resnets)
--coeff: Coefficient for spectral normalization
-mod: whether to use architectural modifications (leaky ReLU + average pooling in skip connections)
--save-path: path/for/saving/model/
As an example, in order to train a Wide-ResNet-28-10 with spectral normalization and architectural modifications on CIFAR-10, use the following:
python train.py \
--seed 1 \
--dataset cifar10 \
--model wide_resnet \
-sn -mod \
--coeff 3.0
Similarly, to train a ResNet-18 with spectral normalization on Dirty-MNIST, use:
python train.py \
--seed 1 \
--dataset dirty-mnist \
--dataset-root /home/user/amnist/ \
--model resnet18 \
-sn \
--coeff 3.0
To evaluate trained models, use evaluate.py. This script can evaluate and aggregate results over multiple experimental runs. For example, if the pretrained models are stored in a directory path /home/user/models, store them using the following directory structure:
models
├── Run1
│ └── wide_resnet_1_350.model
├── Run2
│ └── wide_resnet_2_350.model
├── Run3
│ └── wide_resnet_3_350.model
├── Run4
│ └── wide_resnet_4_350.model
└── Run5
└── wide_resnet_5_350.model
For an ensemble of models, store the models using the following directory structure:
model_ensemble
├── Run1
│ ├── wide_resnet_1_350.model
│ ├── wide_resnet_2_350.model
│ ├── wide_resnet_3_350.model
│ ├── wide_resnet_4_350.model
│ └── wide_resnet_5_350.model
├── Run2
│ ├── wide_resnet_10_350.model
│ ├── wide_resnet_6_350.model
│ ├── wide_resnet_7_350.model
│ ├── wide_resnet_8_350.model
│ └── wide_resnet_9_350.model
├── Run3
│ ├── wide_resnet_11_350.model
│ ├── wide_resnet_12_350.model
│ ├── wide_resnet_13_350.model
│ ├── wide_resnet_14_350.model
│ └── wide_resnet_15_350.model
├── Run4
│ ├── wide_resnet_16_350.model
│ ├── wide_resnet_17_350.model
│ ├── wide_resnet_18_350.model
│ ├── wide_resnet_19_350.model
│ └── wide_resnet_20_350.model
└── Run5
├── wide_resnet_21_350.model
├── wide_resnet_22_350.model
├── wide_resnet_23_350.model
├── wide_resnet_24_350.model
└── wide_resnet_25_350.model
Following are the main parameters for evaluation:
--seed: seed used for initializing the first trained model
--dataset: dataset used for training (cifar10/cifar100)
--ood_dataset: OoD dataset to compute AUROC
--load-path: /path/to/pretrained/models/
--model: model architecture to load (wide_resnet/vgg16)
--runs: number of experimental runs
-sn: whether the model was trained using spectral normalization
--coeff: Coefficient for spectral normalization
-mod: whether the model was trained using architectural modifications
--ensemble: number of models in the ensemble
--model-type: type of model to load for evaluation (softmax/ensemble/gmm)
As an example, in order to evaluate a Wide-ResNet-28-10 with spectral normalization and architectural modifications on CIFAR-10 with OoD dataset as SVHN, use the following:
python evaluate.py \
--seed 1 \
--dataset cifar10 \
--ood_dataset svhn \
--load-path /path/to/pretrained/models/ \
--model wide_resnet \
--runs 5 \
-sn -mod \
--coeff 3.0 \
--model-type softmax
Similarly, to evaluate the above model using feature density, set --model-type gmm. The evaluation script assumes that the seeds of models trained in consecutive runs differ by 1. The script stores the results in a json file with the following structure:
{
"mean": {
"accuracy": mean accuracy,
"ece": mean ECE,
"m1_auroc": mean AUROC using log density / MI for ensembles,
"m1_auprc": mean AUPRC using log density / MI for ensembles,
"m2_auroc": mean AUROC using entropy / PE for ensembles,
"m2_auprc": mean AUPRC using entropy / PE for ensembles,
"t_ece": mean ECE (post temp scaling)
"t_m1_auroc": mean AUROC using log density / MI for ensembles (post temp scaling),
"t_m1_auprc": mean AUPRC using log density / MI for ensembles (post temp scaling),
"t_m2_auroc": mean AUROC using entropy / PE for ensembles (post temp scaling),
"t_m2_auprc": mean AUPRC using entropy / PE for ensembles (post temp scaling)
},
"std": {
"accuracy": std error accuracy,
"ece": std error ECE,
"m1_auroc": std error AUROC using log density / MI for ensembles,
"m1_auprc": std error AUPRC using log density / MI for ensembles,
"m2_auroc": std error AUROC using entropy / PE for ensembles,
"m2_auprc": std error AUPRC using entropy / PE for ensembles,
"t_ece": std error ECE (post temp scaling),
"t_m1_auroc": std error AUROC using log density / MI for ensembles (post temp scaling),
"t_m1_auprc": std error AUPRC using log density / MI for ensembles (post temp scaling),
"t_m2_auroc": std error AUROC using entropy / PE for ensembles (post temp scaling),
"t_m2_auprc": std error AUPRC using entropy / PE for ensembles (post temp scaling)
},
"values": {
"accuracy": accuracy list,
"ece": ece list,
"m1_auroc": AUROC list using log density / MI for ensembles,
"m2_auroc": AUROC list using entropy / PE for ensembles,
"t_ece": ece list (post temp scaling),
"t_m1_auroc": AUROC list using log density / MI for ensembles (post temp scaling),
"t_m1_auprc": AUPRC list using log density / MI for ensembles (post temp scaling),
"t_m2_auroc": AUROC list using entropy / PE for ensembles (post temp scaling),
"t_m2_auprc": AUPRC list using entropy / PE for ensembles (post temp scaling)
},
"info": {dictionary of args}
}
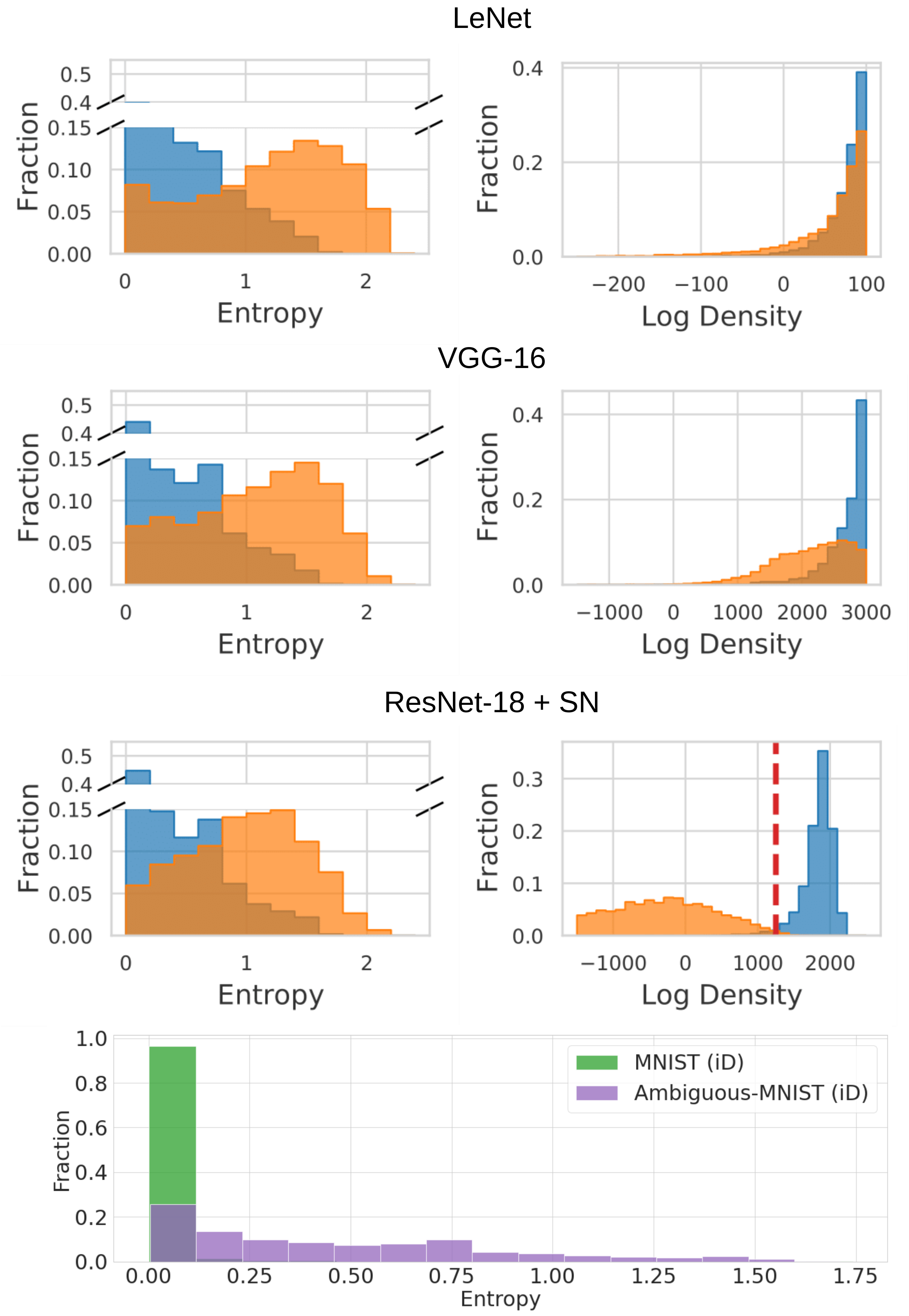
To visualise DDU's performance on Dirty-MNIST (i.e., Fig. 1 of the paper), use fig_1_plot.ipynb. The notebook requires a pretrained LeNet, VGG-16 and ResNet-18 with spectral normalization trained on Dirty-MNIST and visualises the softmax entropy and feature density for Dirty-MNIST (iD) samples vs Fashion-MNIST (OoD) samples. The notebook also visualises the softmax entropies of MNIST vs Ambiguous-MNIST samples for the ResNet-18+SN model (Fig. 2 of the paper). The following figure shows the output of the notebook for the LeNet, VGG-16 and ResNet18+SN model we trained on Dirty-MNIST.
The following table presents results for a Wide-ResNet-28-10 architecture trained on CIFAR-10 with SVHN as the OoD dataset. For the full set of results, refer to the paper.
| Method | Aleatoric Uncertainty | Epistemic Uncertainty | Test Accuracy | Test ECE | AUROC |
|---|---|---|---|---|---|
| Softmax | Softmax Entropy | Softmax Entropy | 95.98+-0.02 | 0.85+-0.02 | 94.44+-0.43 |
| Energy-based | Softmax Entropy | Softmax Density | 95.98+-0.02 | 0.85+-0.02 | 94.56+-0.51 |
| 5-Ensemble | Predictive Entropy | Predictive Entropy | 96.59+-0.02 | 0.76+-0.03 | 97.73+-0.31 |
| DDU (ours) | Softmax Entropy | GMM Density | 95.97+-0.03 | 0.85+-0.04 | 98.09+-0.10 |
To run active learning experiments, use active_learning_script.py. You can run active learning experiments on both MNIST as well as Dirty-MNIST. When running with Dirty-MNIST, you will need to provide a pretrained model on Dirty-MNIST to distinguish between clean MNIST and Ambiguous-MNIST samples. The following are the main command line arguments for active_learning_script.py.
--seed: seed used for initializing the first model (later experimental runs will have seeds incremented by 1)
--model: model architecture to train (resnet18)
-ambiguous: whether to use ambiguous MNIST during training. If this is set to True, the models will be trained on Dirty-MNIST, otherwise they will train on MNIST.
--dataset-root: /path/to/amnist_labels.pt and amnist_samples.pt/
--trained-model: model architecture of pretrained model to distinguish clean and ambiguous MNIST samples
-tsn: if pretrained model has been trained using spectral normalization
--tcoeff: coefficient of spectral normalization used on pretrained model
-tmod: if pretrained model has been trained using architectural modifications (leaky ReLU and average pooling on skip connections)
--saved-model-path: /path/to/saved/pretrained/model/
--saved-model-name: name of the saved pretrained model file
--threshold: Threshold of softmax entropy to decide if a sample is ambiguous (samples having higher softmax entropy than threshold will be considered ambiguous)
--subsample: number of clean MNIST samples to use to subsample clean MNIST
-sn: whether to use spectral normalization during training
--coeff: coefficient of spectral normalization during training
-mod: whether to use architectural modifications (leaky ReLU and average pooling on skip connections) during training
--al-type: type of active learning acquisition model (softmax/ensemble/gmm)
-mi: whether to use mutual information for ensemble al-type
--num-initial-samples: number of initial samples in the training set
--max-training-samples: maximum number of training samples
--acquisition-batch-size: batch size for each acquisition step
As an example, to run the active learning experiment on MNIST using the DDU method, use:
python active_learning_script.py \
--seed 1 \
--model resnet18 \
-sn -mod \
--al-type gmm
Similarly, to run the active learning experiment on Dirty-MNIST using the DDU baseline, with a pretrained ResNet-18 with SN to distinguish clean and ambiguous MNIST samples, use the following:
python active_learning_script.py \
--seed 1 \
--model resnet18 \
-sn -mod \
-ambiguous \
--dataset-root /home/user/amnist/ \
--trained-model resnet18 \
-tsn \
--saved-model-path /path/to/pretrained/model \
--saved-model-name resnet18_sn_3.0_1_350.model \
--threshold 1.0 \
--subsample 1000 \
--al-type gmm
The active learning script stores all results in json files. The MNIST test set accuracy is stored in a json file with the following structure:
{
"experiment run": list of MNIST test set accuracies one per acquisition step
}
When using ambiguous samples in the pool set, the script also stores the fraction of ambiguous samples acquired in each step in the following json:
{
"experiment run": list of fractions of ambiguous samples in the acquired training set
}
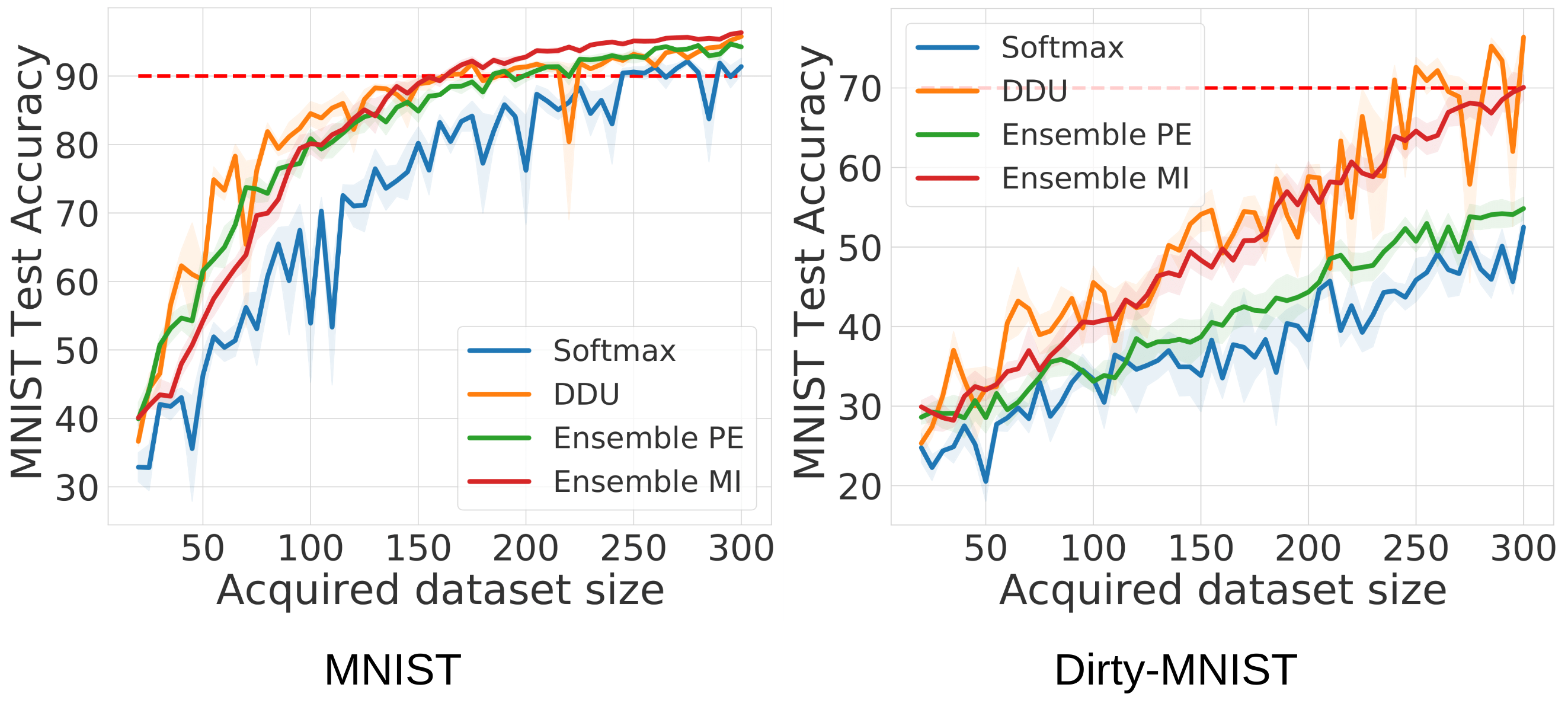
To visualise results from the above json files, use the al_plot.ipynb notebook. The following diagram shows the performance of different baselines (softmax, ensemble PE, ensemble MI and DDU) on MNIST and Dirty-MNIST.
For any questions, please feel free to raise an issue or email us directly. Our emails can be found on the paper.