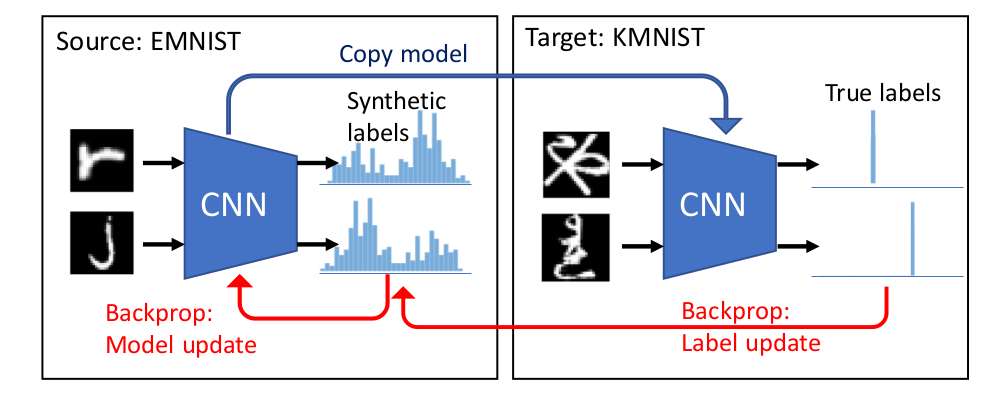
We study the problem of dataset distillation – creating a small set of synthetic examples capable of training a good model. In particular, we study the problem of label distillation – creating synthetic labels for a small set of real images, and show it to be more effective than the prior image-based approach to dataset distillation. Methodologically, we introduce a more robust and flexible meta-learning algorithm for distillation, as well as an effective first-order strategy based on convex optimization layers. Distilling labels with our new algorithm leads to improved results over prior image-based distillation. More importantly, it leads to clear improvements in flexibility of the distilled dataset in terms of compatibility with offthe-shelf optimizers and diverse neural architectures. Interestingly, label distillation can also be applied across datasets, for example enabling learning Japanese character recognition by training only on synthetically labeled English letters.
- Python 3
- CPU or NVIDIA GPU + CUDA
torch >= 1.0.1torchvision >= 0.2.2numpytqdm
Earlier versions may be compatible too (and newer versions may need some changes in torch syntax to be compatible).
The code will automatically download most datasets, but CUB and Kuzushiji-49 need to be downloaded separately.
Instructions for downloading CUB images are here, while instructions for Kuzushiji-49 are here.
After downloading the datasets, place them into data directory so that CUB images are available in data/CUB_200_2011/images (after extracting) and Kuzushiji-49 .npz files are available in data directory.
The easiest way to run experiments is by using one of the configuration json files (modifying parts as needed). Example is shown here, but you can easily use another configuration file (see experiment_configs directory). Description of what the individual parts of the configuration file name mean are in NameDescription.md file located in experiment_configs directory. File configs.txt located in the same directory lists all attached configuration files. We provide configuration files for all main experiments as well as most analysis experiments.
Label distillation experiment with second-order approach:
python label_distillation_or2.py --filepath_to_arguments_json_file experiment_configs/ld_100be_50ib_mn_to_mn_bl_cnn_400e_or2_1234s_v0.json
Label distillation experiment with first-order ridge regression approach:
python label_distillation_rr.py --filepath_to_arguments_json_file experiment_configs/ld_100be_50ib_mn_to_mn_bl_cnn_400e_rr_1234s_v0.json
These scripts do both training and evaluation. If you want to quickly try an experiment without creating a new configuration file (or using one of the provided files), you can use short_experiment.sh.
To make creating configuration files easier, we have created a Jupyter notebook that allows creating many of them efficiently (GenerateExperiments.ipynb in notebooks directory).
You can also specify the arguments directly (for a list of arguments look at arg_extractor.py).
In addition, we provide code for distilling images (data_distillation_or2.py and data_distillation_rr.py). We have not obtained strong results with these, but they can be useful nevertheless.
The results of experiments as well as statistics about the training are saved as json files in results directory, using the name of the experiment. The information stored typically includes information about training and validation error rates across different epochs, test errors across 20 repetitions, total training time (in seconds), best synthetic labels for a set of base examples (original base example labels are stored too) as well as the number of steps used for training validation and test models. Our experiments report error rates rather than accuracies, so to obtain accuracies it is needed to calculate them from the error rates.
If you find this useful for your research, please consider citing:
@article{bohdal2020flexible,
title={Flexible Dataset Distillation: Learn Labels Instead of Images},
author={Bohdal, Ondrej and Yang, Yongxin and Hospedales, Timothy},
journal={arXiv preprint arXiv:2006.08572},
year={2020}
}
This work was supported in part by the EPSRC Centre for Doctoral Training in Data Science, funded by the UK Engineering and Physical Sciences Research Council (grant EP/L016427/1) and the University of Edinburgh.