The goal of baselines is to share code for calculating distance from baselines in ordination space.
You can install this package from github! First you need to have
devtools installed. If not, run install.packages("devtools"), before
running the code below.
# install.packages("devtools")
devtools::install_github("orb16/baselines")library(baselines, quietly = TRUE)
#> rgeos version: 0.5-1, (SVN revision 614)
#> GEOS runtime version: 3.7.2-CAPI-1.11.2
#> Linking to sp version: 1.3-1
#> Polygon checking: TRUE
#> This is vegan 2.5-6
library(vegan, quietly = TRUE)
data("mite")
data("mite.env")
met <- metaMDS(mite, "jaccard")
dlist <- calcEllipseDists(metadf = mite.env, ord = met,
group = "Topo", reflev = "Hummock")
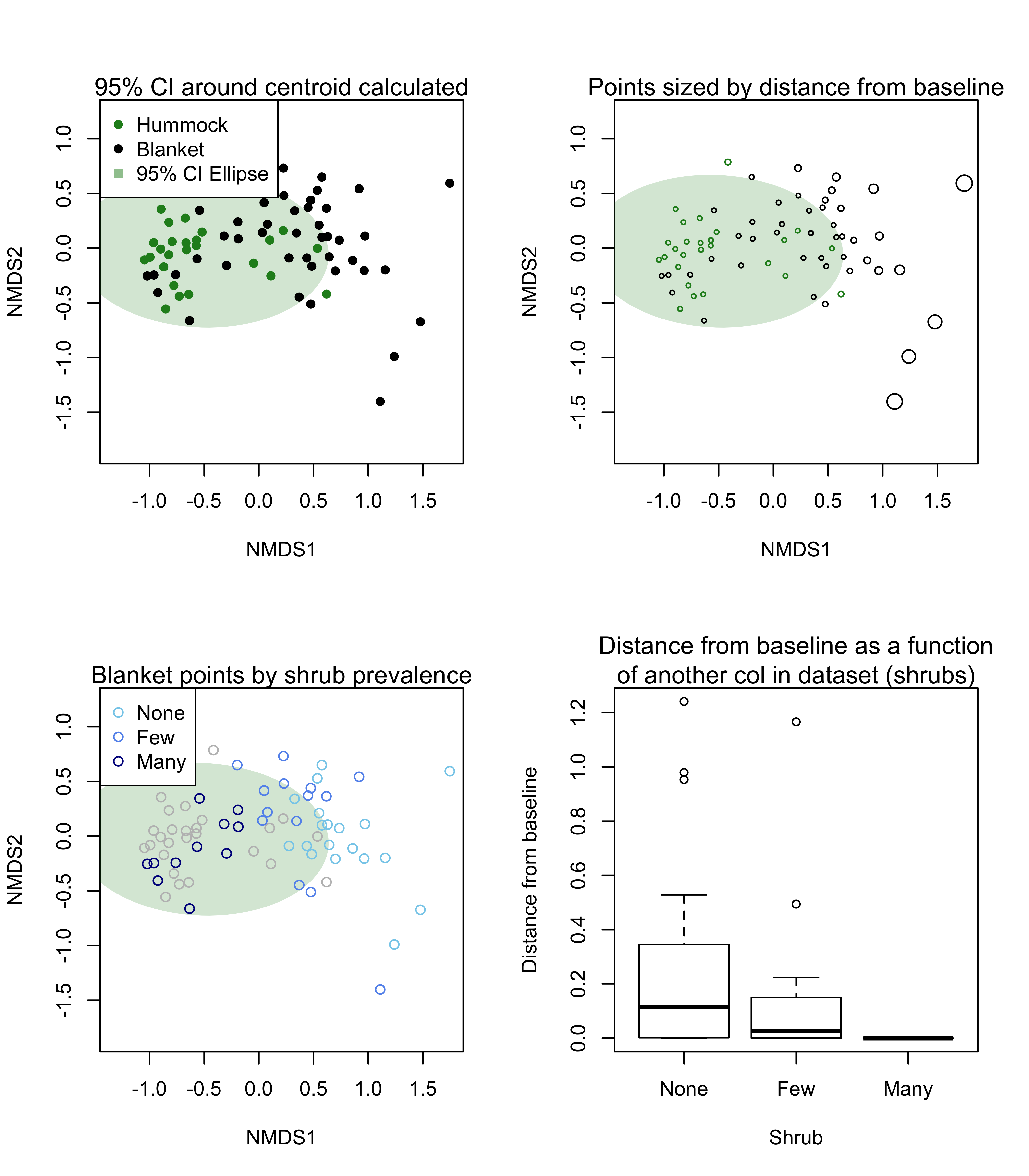
par(mfrow = c(2, 2))
plot(met, type = "n")
plot(dlist[["baseline_polygon"]], add = TRUE,
col = adjustcolor("forestgreen", 0.2),
border = NA)
points(dlist[["all_points"]][dlist[["all_points"]]$Topo == "Hummock", ], col = "forestgreen",
pch = 16)
points(dlist[["all_points"]][dlist[["all_points"]]$Topo == "Blanket", ], col = "black",
pch = 16)
legend("topleft", pch = c(16, 16, 15), col = c("forestgreen", "black", adjustcolor("forestgreen", 0.5)),
legend = c("Hummock", "Blanket", "95% CI Ellipse"))
mtext(side = 3, "95% CI around centroid calculated")
plot(met, type = "n")
plot(dlist[["baseline_polygon"]], add = TRUE,
col = adjustcolor("forestgreen", 0.2),
border = NA)
mtext(side = 3, "Points sized by distance from baseline")
with(dlist[["distDF"]][dlist[["distDF"]]$Topo == "Hummock", ],
points(x = NMDS1, y = NMDS2, cex = distEllipse + 0.5, col = "forestgreen"))
with(dlist[["distDF"]][dlist[["distDF"]]$Topo == "Blanket", ],
points(x = NMDS1, y = NMDS2, cex = distEllipse + 0.5, col = "black"))
plot(met, type = "n")
plot(dlist[["baseline_polygon"]], add = TRUE,
col = adjustcolor("forestgreen", 0.2),
border = NA)
mtext(side = 3, "Blanket points by shrub prevalence")
colDF <- data.frame(Shrub = c("None", "Few", "Many"),
cols = I(c("skyblue", "cornflowerblue", "darkblue")))
with(dlist[["distDF"]][dlist[["distDF"]]$Topo == "Hummock", ],
points(x = NMDS1, y = NMDS2, col = "grey"))
with(dlist[["distDF"]][dlist[["distDF"]]$Topo == "Blanket", ],
points(x = NMDS1, y = NMDS2, col = colDF[match(Shrub, colDF$Shrub), "cols"]))
legend("topleft", pch = 1, legend = colDF$Shrub, col = colDF$cols)
with(dlist[["distDF"]][dlist[["distDF"]]$Topo == "Blanket", ],
plot(x = Shrub, y = distEllipse, xlab = "Shrub",
ylab = "Distance from baseline"))
mtext(side = 3, "Distance from baseline as a function\nof another col in dataset (shrubs)")