minst-dataset
The MNIST of music instrument sounds.
What's going on here?
The goal of this project is to consolidate various disparate solo instrument collections into one big, normalized dataset for ease of use, namely with machine learning in mind. Simply put, this aims to be the MNIST for music audio processing.
Installing Dependencies
$ pip install git+git://github.com/ejhumphrey/minst-dataset.git
$ cd minst-dataset
$ pip install -U -r requirements.txt
OR
$ make deps
Testing your Install / Setup
This project ships with just enough data to test itself.
$ make test
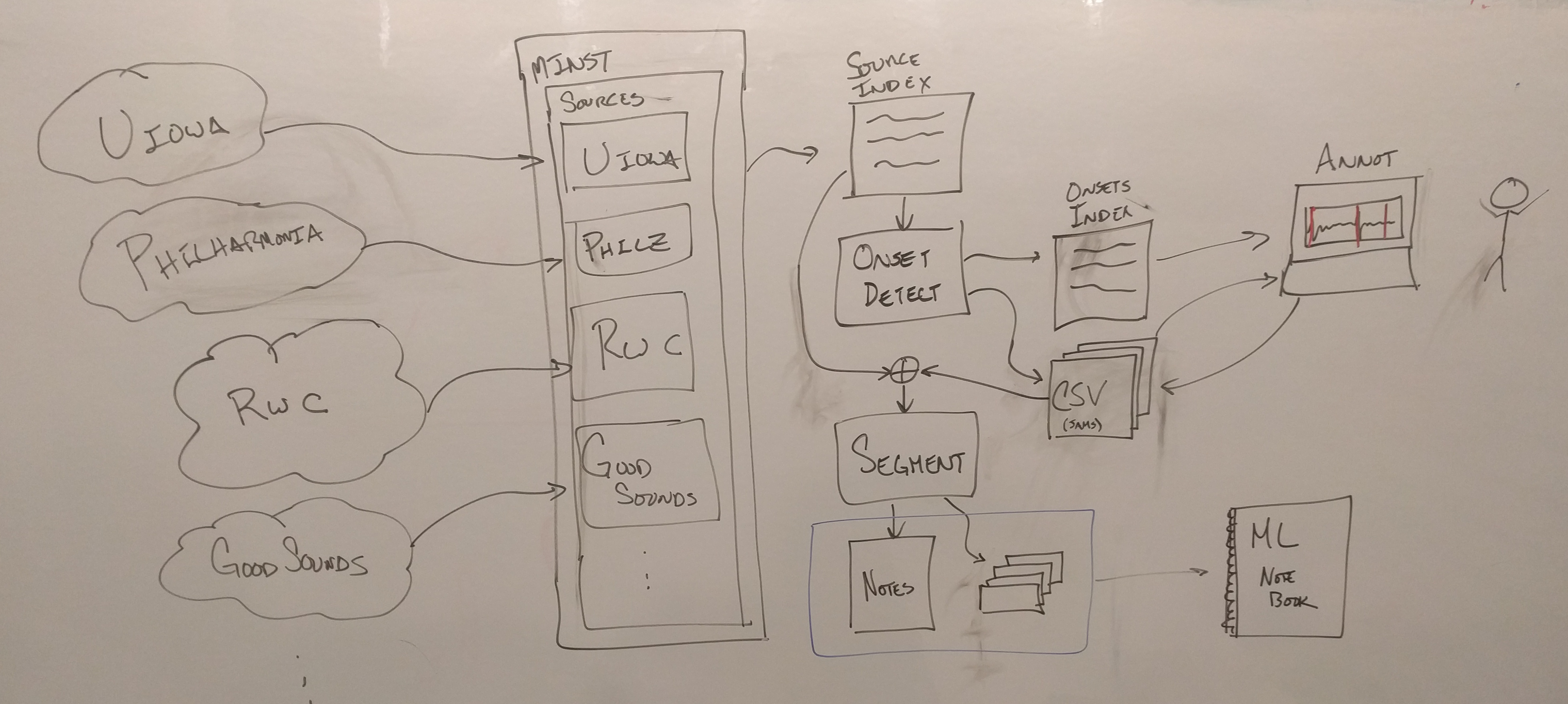
Building the dataset from scratch
Directory Structure
This library expects a certain directory structure for everything to work nicely. The following is a flyover of what this should look like locally by default (according to Makefile):
{DATA_DIR}/
uiowa/
...
RWC Instruments/
...
philharmonia/
...
The only value in the Makefile you may need to update is DATA_DIR, in the case that you would prefer the data live elsewhere when downloaded, e.g. a different hard drive.
Get the data
This project uses four different solo instrument datasets.
We provide "manifest" files with which one can download the first two collections. For access to the third (RWC), you should contact the kind folks at AIST. Access to Good-Sounds is public, but requires that you fill out a form first.
To download the available data, you can invoke the following from your cloned repository:
$ make download
(which is equivalent to:)
$ python scripts/download.py data/uiowa.json ~/data/uiowa
...
$ python scripts/download.py data/philharmonia.json ~/data/philharmonia
Preparing note audio using annotated onsets
Assuming your data is downloaded and available, you can use the following to build the index from the downloaded files, and then extract the note audio from it.
Warning: extracting notes could take up to an hour with all four datasets.
$ make build
Afterwards, the annotated notes should be available in these files:
uiowa_notes.csv
phil_notes.csv
rwc_notes.csv
goodsounds_notes.csv
If the dataset is not available on your machine, make build should skip it.
Build final dataset *.csv's for experimenting
$ make dataset
This will generate the following files in your data_dir (~/data/minst):
-
master_index.csv A master dataset file containing pointers to the audio and targets/metadata. The "index" is the first column.
-
rwc_partitions.csv
-
uiowa_partitions.csv
-
philharmonia_partitions.csv These csvs designed to be sort of "three-fold" cross validation, where each dataset acts as a hold-out test set. The dataset in the name is the test set.
These csvs contain two columns: "index", and "partition".
"Index" is the index in master_index.csv, and "partition" is one of ['train', 'val', 'test'], and specifies which dataset partition to use
the files for.
If you are loading using python/pandas, getting the training data would look like this:
master_df = pd.read_csv('master_index.csv', index_col=0)
partition_df = pd.read_csv('rwc_partitions.csv')
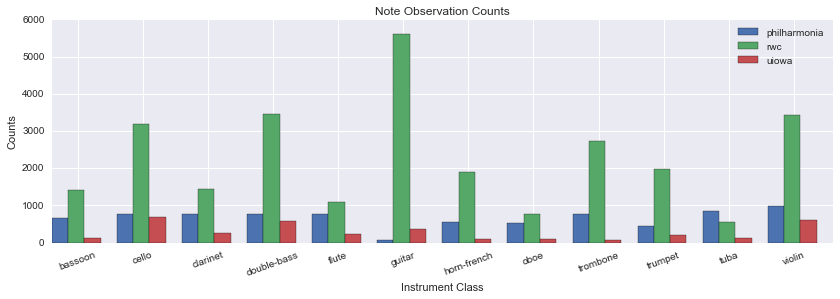
train_df = master_df.loc[(partition_df['partition'] == 'train').index]Note Counts Per Dataset for Accepted Instruments
| Instrument | UIowa | Philharmonia | RWC | Good-Sounds |
|---|---|---|---|---|
| totals | 3417 | 7923 | 27557 | 12015 |
| bassoon | 122 | 648 | 1405 | |
| cello | 681 | 776 | 3196 | 2118 |
| clarinet | 258 | 770 | 1433 | 3359 |
| double-bass | 587 | 781 | 3465 | |
| flute | 227 | 781 | 1095 | 2308 |
| guitar | 352 | 71 | 5618 | |
| horn-french | 96 | 546 | 1896 | |
| oboe | 104 | 539 | 770 | 494 |
| trombone | 66 | 769 | 2738 | |
| trumpet | 212 | 433 | 1965 | 1883 |
| tuba | 111 | 838 | 540 | |
| violin | 601 | 971 | 3436 | 1853 |
Appendix: Computing / finding note onsets
This repository contains some generated / annotated onsets for the instruments we have selected in the taxonomy. If you wish to annotate more, however, you will need to do the following:
Both the UIowa and RWC collections ship as recordings with multiple notes per file. To establish a clean dataset with which to work, it is advisable to split up these recordings (roughly) at note onsets.
Somewhat surprisingly, modern onset detection algorithms have not been optimized for this use case, and it is challenging to find a single parameterization that works well over the entire collection. To overcome this deficiency, we take a human-in-the-loop approach to robustly arrive at good cut-points for these (and future) collections.
First, collect a single dataset:
$ make uiowa_index.csv
OR
$ python scripts/collect_data.py uiowa path/to/download uiowa_index.csv
Optionally, you can run a segmentation algorithm over the resulting index. This will generate a number of best-guess onsets, saved out as CSV files under the same index as the collection, and an new dataframe tracking where these aligned files live locally (uiowa_onsets/segment_index.csv below):
Warning: buggy.
$ python scripts/compute_note_onsets.py uiowa_index.csv uiowa_onsets \
segment_index.csv --mode logcqt --num_cpus -1 --verbose 50
Either way, you'll want to verify and correct (as needed) the estimated onsets. To do so, drop into the annotation routine by the following:
$ python scripts/annotate.py uiowa_onsets/segment_index.csv
GUI Command Summary:
spacebar: add / remove a marker at the location of the mouse cursorup arrow: move all markers .1s to the leftdown arrow: move all markers .1s to the rightleft arrow: move all markers .01s to the leftright arrow: move all markers .01s to the rightd: delete marker within 1s of cursorD: delete marker within 5s of cursor1: Replace all markers with envelope_onsets(wait=.008s)2: Replace all markers with envelope_onsets(wait=.01s)3: Replace all markers with envelope_onsets(wait=.02s)4: Replace all markers with envelope_onsets(wait=.05s)6: Replace all markers with logcqt_onsets(wait=.01s)7: Replace all markers with logcqt_onsets(wait=.02s)wwill write the current markersqwill close current file without savingxwill write onset data and closeQTo kill the process entirely
Improvements / enhancements are more than welcomed.