MTDS applied to video data of a double pendulum. This is code for the updated MTDS project -- for the previous version of the paper see https://arxiv.org/abs/1910.05026. The details for the double pendulum work are only available in the updated paper which is currently under review. This repo is therefore not yet properly packaged up for an end user, although it will not take much extra work. For those who are interested, the code is fairly well annotated, and there's some commands to get you started below.
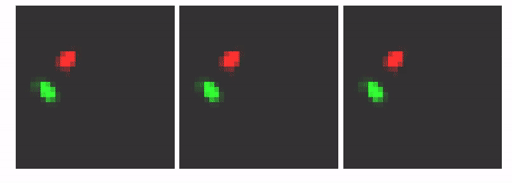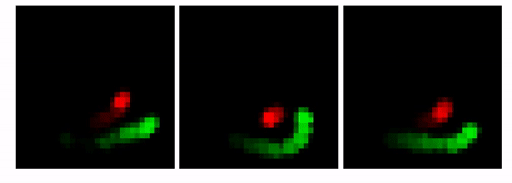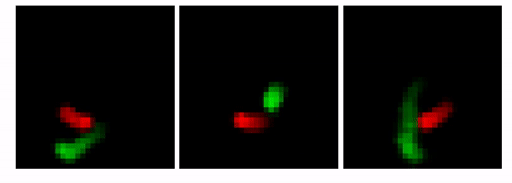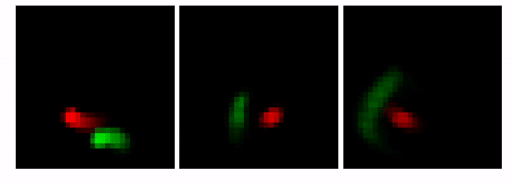
Our goal here is to show that by changing the architecture to an MTDS construct, we can permit customizable predictions from a seq2seq RNN (with convolutional encoder/decoder). An example is shown below. Here, the prediction for the first 10 frames is identical (shown with the lighter grey), after which we adjust the latent variable to yield three possible predictions.
For more examples, with some commentary, see https://sites.google.com/view/mtds-customized-predictions/home.
In this example, we load a pre-trained model (please download the data repository from https://gin.g-node.org/alxbird/dblpendulum -- note
that the CLI appears not to work for downloading this file, at least if you're not logged into gin, so just download it directly from the webpage. Unpack the data into a folder called
data within the structure of this repo. (YAML files in saved_models will point here.)
using Flux
# Load MTDS libraries
include("src/modelutils.jl")
include("src/datagen.jl")
include("src/seq2seq.jl")
include("src/multitask.jl")
const unsqueeze = modelutils.unsqueeze; # trivial but useful function
const chan3cat = modelutils.chan3cat # concatenate with (zero) 3rd colour channel
standardize(x; dims=1) = (x .- mean(x; dims=dims)) ./ std(x; dims=dims);
make_untracked(x) = mapleaves(Tracker.data, x) # remove ADdata_xy, data_θ, data_meta = datagen.generate_data(); # _seed argument is fixed, but can be changed
# data_xy - (x,y) co-ordinates of bob1 and bob2.
# data_θ - (θ, θdot) state of bob1 and bob2 (for reference, but unused in model).
# data_meta - metadata about each sequence incl. ODE parameters and initial conditions.
# To reduce memory usage, we won't "image" all of these co-ordinates for the videos
# but instead construct these "on-demand". Julia can do this in microseconds.
constr_image(x) = (@assert length(x) == 4; datagen.gen_pic_circ2_2chan_cood_tf(x[1:2], x[3:4]))
tT = 80
cseq = [unsqueeze(constr_image(data_xy[:test][10][tt, :]), 4) for tt in 1:tT] # |> gpu # for loading to GPU.video_mtgru_pred_fixb_u = mtmodel.load_model_from_def("saved_models/mtgru_video_fixb_pred_300.yml")
video_mtgru_pred_fixb_u = make_untracked(video_mtgru_pred_fixb_u) # |> gpu # for loading to GPU.# Predict forward `T_steps` from the first `T_enc` values of the current sequence (`cseq`).
# Note that the prediction is in logits, and we must transform to [0,1] via sigmoid.
logit_yhat = video_mtgru_pred_fixb_u(cseq; T_enc=20, T_steps=80)
yhat = map(x->σ.(x), logit_yhat); # perform (elementwise) sigmoid transform to listWe extract all of the initial x0 variables for each sequence in the test set. (This is quick, and amortizes the cost incurred otherwise from each sample of z. The code is similar to above, but see ?mtmodel.posterior_samples for more details.
# initial x0 of test set
m = video_mtgru_pred_fixb_u
testx0 = []
for i = 1:10
cbatch = (10*(i-1)+1):10*i
cseqs = Flux.batch([data_xy[:test][s][1:10,:] for s in cbatch])
cseqs = [cat([datagen.gen_pic_circ2_2chan_cood_tf(cseqs[tt, 1:2, s], cseqs[tt, 3:4, s])
for s in 1:10]..., dims=4) for tt in 1:10]; # [32×32×2×nbatch for t in 1:70].
post = mtmodel.posterior_samples(m, cseqs, []; z=0, c=0, T_enc=20)
push!(testx0, (post[2][1], post[3][1]))
end
testx0 = (reduce(hcat, [x[1] for x in testx0]), reduce(hcat, [x[2] for x in testx0]))
testx0post = [MvNormal(testx0[1][:,i], Diagonal(testx0[2][:,i].^2)) for i in 1:100];This is not essential, but tidies up the results later.
using NNlib
struct MCPosterior
samples::AbstractMatrix
logW::Vector
ess::AbstractFloat
end
weights(P::MCPosterior) = NNlib.softmax(P.logW)
resample(P::MCPosterior, N::Int) = P.samples[rand(Categorical(weights(P)), N),:]
Base.length(P::MCPosterior) = length(P.logW)
ess(P::MCPosterior) = P.ess
struct GMM{T <: Real}
pis::Vector{T}
mus::AbstractMatrix{T}
covs::AbstractArray{T}
endThese are calculated based on the aggregate posterior of the training set, but we take a shortcut here:
pzmean, pzcov = Float32[-0.012118647, 0.18318911], Float32[0.187806 0.0241003; 0.0241003 0.230048];
p_z = MvNormal(pzmean, pzcov)function generate_logjoint(m, test_ix, to_time, data_xy, p_z; batch_size=15, β_def=1f0)
x0 = trainx0post[test_ix].μ
function logjoint(S, β=β_def)
# β for annealing
bsz = batch_size
n, d = size(S)
@argcheck d == 2
y = data_xy[tvt][test_ix][1:to_time,:]
y = [datagen.gen_pic_circ2_2chan_cood_tf(y[tt, 1:2], y[tt, 3:4])
for tt in 1:to_time]; # [32×32×2 for t in 1:to_time].
Z = gpu(S[:, 1:2]')
nllh = map(Iterators.partition(1:n, bsz)) do cbatch
x0_init = gpu(repeat(x0, 1, length(cbatch))) # using mean posterior x0
txb = mtmodel.online_inference_BCE(m, x0_init, Z[:, cbatch], y)
cpu(dropdims(sum(txb, dims=1), dims=1)) # online BCE result is T × batchsize; sum over time
end
llh = - reduce(vcat, nllh)
# prior
lp_z = logpdf(p_z, cpu(Z))
return lp_z + β*llh
end
endtest_ix = 5
smp_out = []
dist_out = []
k = 3
nepochs = 3
gmm_smps = 2000
t0 = time()
t_begin = copy(t0)
printfmtln("========= TEST INDEX {:d} ========", test_ix)
S, logW, pis, mus, covs = nothing, nothing, nothing, nothing, nothing # local scope
for t in 5:5:20
IS_tilt, nepochs, c_ess = 1.3f0, 3, 0
if t==5
for j = 1:3
lj = generate_logjoint(video_mtgru_u, test_ix, t, data_xy, p_z, p_c_0; batch_size=10, β_def=1f0)
S, logW, pis, mus, covs = infer.amis(lj, MvNormal(vcat(p_z.μ, p_c_0.μ),
Diagonal(vcat(diag(p_z.Σ), diag(p_c_0.Σ)))), k; nepochs=nepochs, gmm_smps=gmm_smps)
c_ess = infer.eff_ss(softmax(logW))
(c_ess > 100) && break
IS_tilt, nepochs = IS_tilt*1.3, nepochs + 1
println("Retrying...")
end
else
for j in 1:3
lj = generate_logjoint(video_mtgru_u, test_ix, t, data_xy, p_z, p_c_0; batch_size=50, β_def=1f0)
S, logW, pis, mus, covs = infer.amis(lj, pis, mus, covs, S, logW; nepochs=nepochs,
gmm_smps=gmm_smps, IS_tilt=1.3)
c_ess = infer.eff_ss(softmax(logW))
(c_ess > 100) && break
IS_tilt, nepochs = IS_tilt*1.3, nepochs + 1
println("Retrying...")
end
end
push!(smp_out, MCPosterior(S, logW, c_ess))
push!(dist_out, (pis, mus, covs))
printfmtln("Posterior {:d}, ess={:.2f}, time taken = {:.2f}s ({:.2f}).", t, c_ess,
time()-t_begin, time() - t0); flush(stdout);
t0 = time()
end