Nivolumab-treatment-for-r-r-EBV-HLH
All single cell data in this paper was submitted with the scripts.
Citation
Our paper has been published on the Blood PMID 31914172
You could read our paper here, and all the supplemental materials are in here
You could downloaded raw data from GEO Database GSE138504
Single cell Libraries constructed
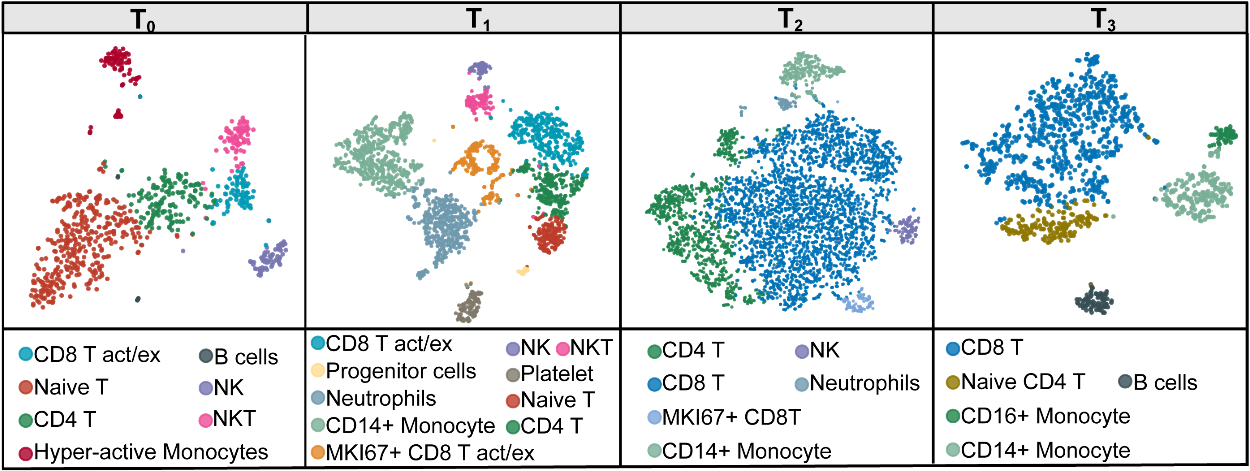
Single cell RNA sequencing (scRNA-seq) was performed with peripheral blood cells before (Day 0, T0), during nivolumab treatment (Day 7, T1; Day 21, T2), and when plasma EBV turned negative (Day 76, T3) in 1 patient (patient 7). scRNA-seq libraries were generated following the recommended protocol of the 3’ scRNA-seq 10X genomics platform and using v2 chemistry, and sequenced data was collected by illumina NovaSeq 6000 sequencing.
The version 2. Basic analysis of all merge data of HLH
To better visualize our data, we had modified a lot of functions from Seurat. If you want to used them, you need to loading them into your R environment.
library(Seurat)
library(ggplot2)
library(cowplot)
source("./scripts/modified_Function_HLH.R")Then, you could begin the data analyze from following codes.
-
To repeat the figure mentioned and used in our paper, the
.rdsfiles could be downloaded from GEO Database GSE138504Then, you could use following codes to make the visualization.
HLH_T1 <- readRDS("./processed_data/anno_HLH_T1.rds")
HLH_T2 <- readRDS("./processed_data/anno_HLH_T2.rds")
HLH_T3 <- readRDS("./processed_data/anno_HLH_T3.rds")
HLH_T4 <- readRDS("./processed_data/anno_HLH_T4.rds")
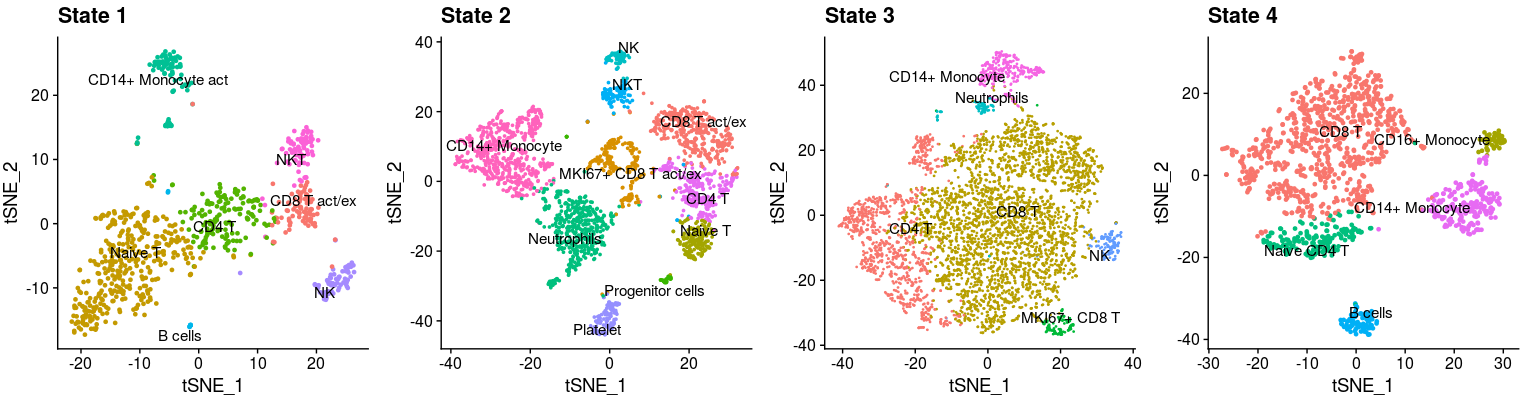
p1 <- DimPlot(object = HLH_T1, reduction = "tsne", label = TRUE,repel=T) +NoLegend()+labs(title="State 1")
p2 <- DimPlot(object = HLH_T2, reduction = "tsne", label = TRUE,repel=T) +NoLegend()+labs(title="State 2")
p3 <- DimPlot(object = HLH_T3, reduction = "tsne", label = TRUE,repel=T) +NoLegend()+labs(title="State 3")
p4 <- DimPlot(object = HLH_T4, reduction = "tsne", label = TRUE,repel=T) +NoLegend()+labs(title="State 4")
plot_grid(p1,p2,p3,p4,nrow=1)- To make more analysis in our HLH scRNA-seq data, we integrated more tools and processing steps for analysis, including integrating multiple samples and better visualization.
Here is the code to merge all the data of HLH
HLH_T1$group <- "T1"
HLH_T2$group <- "T2"
HLH_T3$group <- "T3"
HLH_T4$group <- "T4"
library(reticulate)
library(ReductionWrappers)
library(s2a)
all_data <- merge(x = HLH_T1, y = c(HLH_T2,HLH_T3,HLH_T4))
all_merge <- all_data %>%
Seurat::NormalizeData(verbose = FALSE) %>%
FindVariableFeatures(selection.method = "vst") %>%
ScaleData(verbose = TRUE) %>%
RunPCA(pc.genes = all_merge@var.genes, npcs = 30, verbose = FALSE)
all_merge <- all_merge %>%
RunUMAP(dims = 1:15) %>%
RunTSNE(dims = 1:15) %>%
FindNeighbors(dims = 1:15) %>%
FindClusters(resolution = 0.1) %>%
DoopenTSNE(reduction_use = "pca", reduction_save = "openTSNE",dims_use = 1:15) %>%
DoPHATE(reduction_use = "pca", reduction_save = "phate",dims_use = 1:15) %>%
DoPhenoGraph(reduction_use = "pca", k = 500,prefix = "PhenoGraph")
all_merge <- XY_RunURD_DM(all_merge,assay = "RNA",key = "URDDM",sigma=15,visua_group="group")
all_merge <- DooptSNE(all_merge, reduction_use = "pca", reduction_save = "optsne",dims_use = 1:15)
all_merge <- DoForceAtlas2(all_merge, reduction_use = "pca", reduction_save = "fa2",dims_use = 1:15)
mcsaveRDS(all_merge,"./all/all_merge.rds",mc.cores=20)And the .rds files could be accessed from ./rds_files/all_merge.rds
all_merge <- readRDS("./all/all_merge.rds")
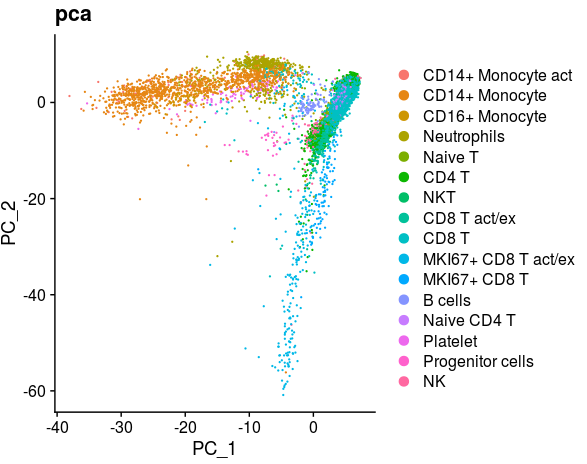
DimPlot(object = all_merge, reduction = "pca",label=FALSE,repel=TRUE,group.by="new_anno")+labs(title="pca")library(scales)
all_merge$new_anno <- factor(all_merge$new_anno,levels=c("CD14+ Monocyte act","CD14+ Monocyte",
"CD16+ Monocyte","Neutrophils","Naive T","CD4 T","NKT",
"CD8 T act/ex","CD8 T","MKI67+ CD8 T act/ex","MKI67+ CD8 T",
"B cells","Naive CD4 T","Platelet","Progenitor cells","NK"))
col_sel <- hue_pal()(length(as.character(levels(all_merge$new_anno))))
col <- col_sel[1:length(as.character(levels(all_merge$new_anno)))]
names(col) <- as.character(levels(all_merge$new_anno))
col <- c(col,"#efefef")
names(col)[length(col)] <- "OTS"
all_merge$new_anno5 <- all_merge$new_anno
all_merge$new_anno5 <- as.character(all_merge$new_anno5)
all_merge$new_anno5[setdiff(1:length(all_merge$new_anno5),c(which(all_merge$new_anno5=="CD16+ Monocyte"),
which(all_merge$new_anno5=="CD14+ Monocyte act"),
which(all_merge$new_anno5=="CD14+ Monocyte"),
which(all_merge$new_anno5=="Neutrophils")
))] <- "OTS"
all_merge$new_anno5 <- factor(all_merge$new_anno5,levels=c("OTS","CD14+ Monocyte","CD16+ Monocyte","Neutrophils","CD14+ Monocyte act"))
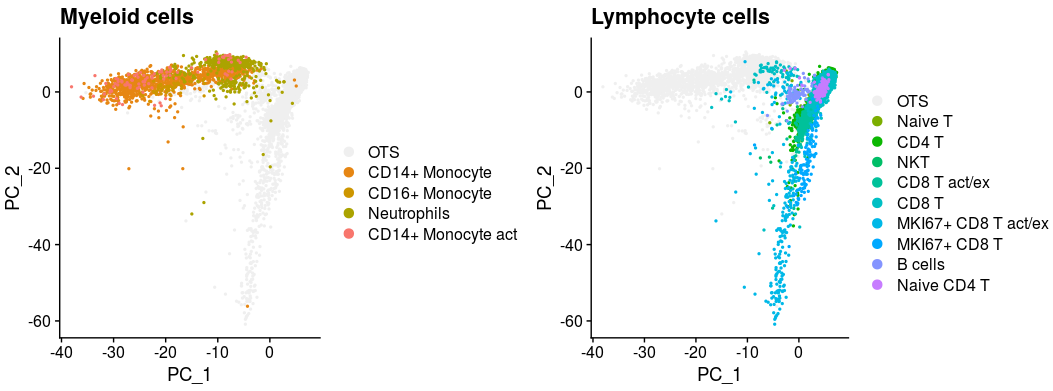
p1 <- XY_DimPlot(all_merge, reduction = 'pca', label = FALSE,repel=TRUE, pt.size = .5,group.by="new_anno5",cols=col[levels(all_merge$new_anno5)]) + labs(title="Myeloid cells")
all_merge$new_anno5 <- all_merge$new_anno
all_merge$new_anno5 <- as.character(all_merge$new_anno5)
all_merge$new_anno5[setdiff(1:length(all_merge$new_anno5),c(which(all_merge$new_anno5=="Naive T"),
which(all_merge$new_anno5=="CD4 T"),
which(all_merge$new_anno5=="NKT"),
which(all_merge$new_anno5=="CD8 T act/ex"),
which(all_merge$new_anno5=="CD8 T"),
which(all_merge$new_anno5=="MKI67+ CD8 T act/ex"),
which(all_merge$new_anno5=="B cells"),
which(all_merge$new_anno5=="Naive CD4 T"),
which(all_merge$new_anno5=="MKI67+ CD8 T")
))] <- "OTS"
all_merge$new_anno5 <- factor(all_merge$new_anno5,levels=c("OTS","Naive T","CD4 T","NKT",
"CD8 T act/ex","CD8 T","MKI67+ CD8 T act/ex","MKI67+ CD8 T","B cells","Naive CD4 T"))
p2 <- XY_DimPlot(all_merge, reduction = 'pca', label = FALSE,repel=TRUE, pt.size = .5,group.by="new_anno5",cols=col[levels(all_merge$new_anno5)]) + labs(title="Lymphocyte cells")
plot_grid(p1,p2,nrow=1)all_merge$new_anno5 <- all_merge$new_anno
all_merge$new_anno5 <- as.character(all_merge$new_anno5)
all_merge$new_anno5[setdiff(1:length(all_merge$new_anno5),c(which(all_merge$new_anno5=="CD14+ Monocyte act")))] <- "OTS"
all_merge$new_anno5 <- factor(all_merge$new_anno5,levels=c("OTS","CD14+ Monocyte act"))
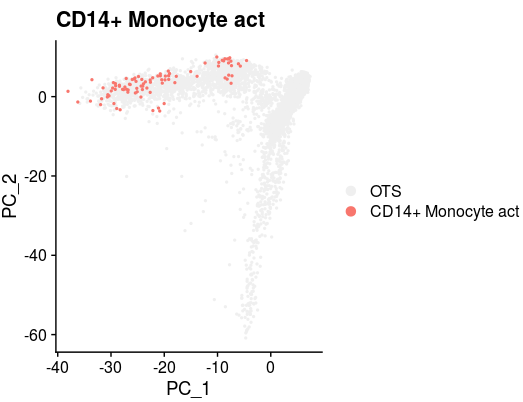
XY_DimPlot(all_merge, reduction = 'pca', label = FALSE,repel=TRUE, pt.size = .5,group.by="new_anno5",cols=col[levels(all_merge$new_anno5)]) + labs(title="CD14+ Monocyte act")all_merge$new_anno5 <- all_merge$new_anno
all_merge$new_anno5 <- as.character(all_merge$new_anno5)
all_merge$new_anno5[setdiff(1:length(all_merge$new_anno5),c(which(all_merge$new_anno5=="CD4 T"),
which(all_merge$new_anno5=="CD8 T act/ex"),
which(all_merge$new_anno5=="MKI67+ CD8 T act/ex"),
which(all_merge$new_anno5=="MKI67+ CD8 T")
))] <- "OTS"
all_merge$new_anno5 <- factor(all_merge$new_anno5,levels=c("OTS","CD4 T","CD8 T act/ex","MKI67+ CD8 T act/ex","MKI67+ CD8 T"))
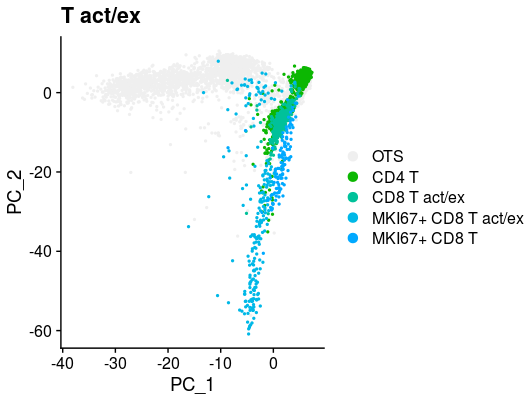
XY_DimPlot(all_merge, reduction = 'pca', label = FALSE,repel=TRUE, pt.size = .5,group.by="new_anno5",cols=col[levels(all_merge$new_anno5)]) + labs(title="T act/ex")all_merge$new_anno5 <- all_merge$new_anno
all_merge$new_anno5 <- as.character(all_merge$new_anno5)
all_merge$new_anno5[which(all_merge$group!="T1")] <- "OTS"
all_merge$new_anno5 <- factor(all_merge$new_anno5,levels=c("OTS","B cells","CD14+ Monocyte act","CD4 T","CD8 T act/ex","Naive T","NKT","NK"))
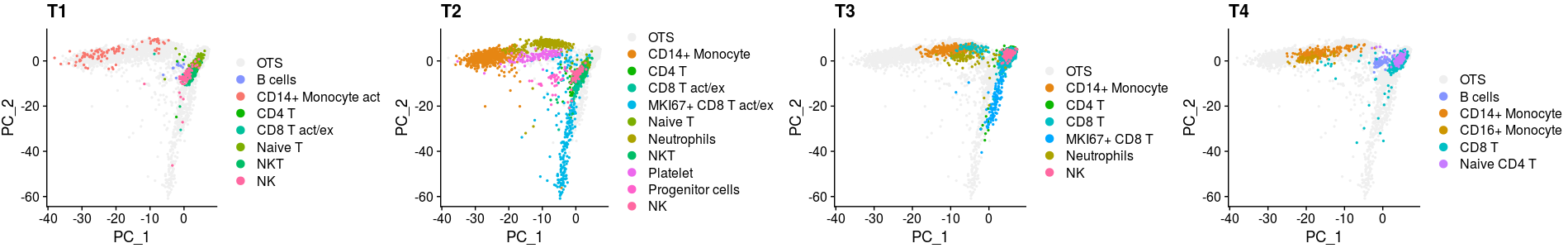
p1 <- XY_DimPlot(all_merge, reduction = 'pca', label = FALSE,repel=TRUE, pt.size = .5,group.by="new_anno5",cols=col[levels(all_merge$new_anno5)]) + labs(title="T1")
all_merge$new_anno5 <- all_merge$new_anno
all_merge$new_anno5 <- as.character(all_merge$new_anno5)
all_merge$new_anno5[which(all_merge$group!="T2")] <- "OTS"
all_merge$new_anno5 <- factor(all_merge$new_anno5,levels=c("OTS","CD14+ Monocyte","CD4 T","CD8 T act/ex","MKI67+ CD8 T act/ex","Naive T","Neutrophils","NKT","Platelet","Progenitor cells","NK"))
p2 <- XY_DimPlot(all_merge, reduction = 'pca', label = FALSE,repel=TRUE, pt.size = .5,group.by="new_anno5",cols=col[levels(all_merge$new_anno5)]) + labs(title="T2")
all_merge$new_anno5 <- all_merge$new_anno
all_merge$new_anno5 <- as.character(all_merge$new_anno5)
all_merge$new_anno5[which(all_merge$group!="T3")] <- "OTS"
all_merge$new_anno5 <- factor(all_merge$new_anno5,levels=c("OTS","CD14+ Monocyte","CD4 T","CD8 T","MKI67+ CD8 T","Neutrophils","NK"))
p3 <- XY_DimPlot(all_merge, reduction = 'pca', label = FALSE,repel=TRUE, pt.size = .5,group.by="new_anno5",cols=col[levels(all_merge$new_anno5)]) + labs(title="T3")
all_merge$new_anno5 <- all_merge$new_anno
all_merge$new_anno5 <- as.character(all_merge$new_anno5)
all_merge$new_anno5[which(all_merge$group!="T4")] <- "OTS"
all_merge$new_anno5 <- factor(all_merge$new_anno5,levels=c("OTS","B cells","CD14+ Monocyte","CD16+ Monocyte","CD8 T","Naive CD4 T"))
p4 <- XY_DimPlot(all_merge, reduction = 'pca', label = FALSE,repel=TRUE, pt.size = .5,group.by="new_anno5",cols=col[levels(all_merge$new_anno5)]) + labs(title="T4")
plot_grid(p1,p2,p3,p4,nrow=1)all_merge$new_anno5 <- all_merge$new_anno
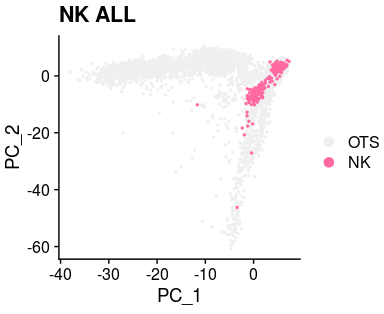
all_merge$new_anno5 <- as.character(all_merge$new_anno5)
all_merge$new_anno5[setdiff(1:length(all_merge$new_anno5),c(which(all_merge$new_anno5=="NK")
))] <- "OTS"
all_merge$new_anno5 <- factor(all_merge$new_anno5,levels=c("OTS","NK"))
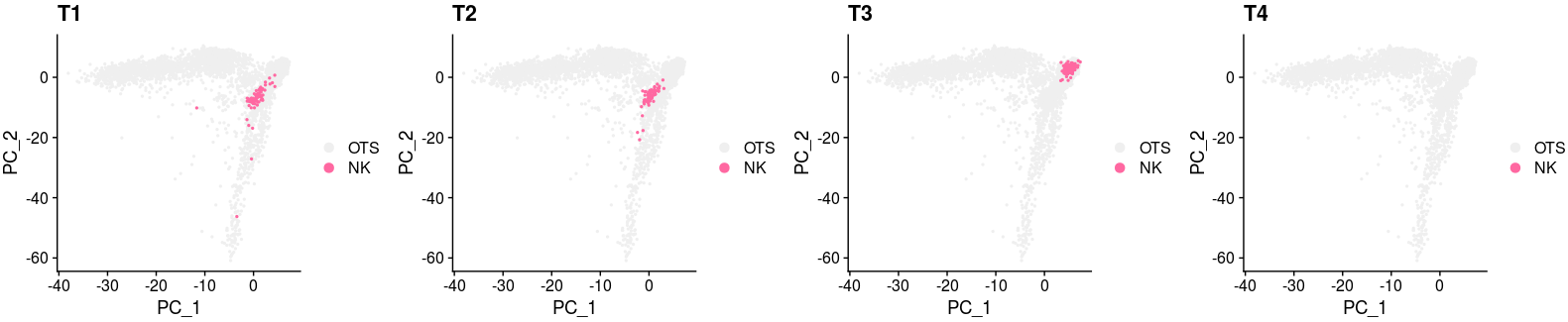
XY_DimPlot(all_merge, reduction = 'pca', label = FALSE,repel=TRUE, pt.size = .5,group.by="new_anno5",cols=col[levels(all_merge$new_anno5)])+ labs(title="NK ALL")all_merge$new_anno6 <- all_merge$new_anno5
all_merge$new_anno6 <- as.character(all_merge$new_anno6)
all_merge$new_anno6[which(all_merge$group!="T1")] <- "OTS"
all_merge$new_anno6 <- factor(all_merge$new_anno6,levels=c("OTS","NK"))
p1 <- XY_DimPlot(all_merge, reduction = 'pca', label = FALSE,repel=TRUE, pt.size = .5,group.by="new_anno6",cols=col[levels(all_merge$new_anno6)]) + labs(title="T1")
all_merge$new_anno6 <- all_merge$new_anno5
all_merge$new_anno6 <- as.character(all_merge$new_anno6)
all_merge$new_anno6[which(all_merge$group!="T2")] <- "OTS"
all_merge$new_anno6 <- factor(all_merge$new_anno6,levels=c("OTS","NK"))
p2 <- XY_DimPlot(all_merge, reduction = 'pca', label = FALSE,repel=TRUE, pt.size = .5,group.by="new_anno6",cols=col[levels(all_merge$new_anno6)]) + labs(title="T2")
all_merge$new_anno6 <- all_merge$new_anno5
all_merge$new_anno6 <- as.character(all_merge$new_anno6)
all_merge$new_anno6[which(all_merge$group!="T3")] <- "OTS"
all_merge$new_anno6 <- factor(all_merge$new_anno6,levels=c("OTS","NK"))
p3 <- XY_DimPlot(all_merge, reduction = 'pca', label = FALSE,repel=TRUE, pt.size = .5,group.by="new_anno6",cols=col[levels(all_merge$new_anno6)]) + labs(title="T3")
all_merge$new_anno6 <- all_merge$new_anno5
all_merge$new_anno6 <- as.character(all_merge$new_anno6)
all_merge$new_anno6[which(all_merge$group!="T4")] <- "OTS"
all_merge$new_anno6 <- factor(all_merge$new_anno6,levels=c("OTS","NK"))
p4 <- XY_DimPlot(all_merge, reduction = 'pca', label = FALSE,repel=TRUE, pt.size = .5,group.by="new_anno6",cols=col[levels(all_merge$new_anno6)]) + labs(title="T4")
plot_grid(p1,p2,p3,p4,nrow=1)The version 1. Overview of submitted data
The .rds files generated by Seurat V3 were submitted. And the annotation information stored in meta.data.
Consideration the limitation of github regulation (each file cannot bigger than 25Mb), so we splited all files in many individual files (10Mb). And after your downloading, you could use the code to merged them and compressed files. The code and example were as following:
cat anno_HLH_T1.zip* > anno_HLH_T1.zip
unzip anno_HLH_T1.zipAnd then you could export the rds.files in R