The goals of this project are threefold: (1) to explore development of a machine learning algorithm to distinguish chest X-rays of individuals with respiratory illness testing positive for COVID-19 from other X-rays, (2) to promote discovery of patterns in such X-rays via machine learning interpretability algorithms, and (3) to build more robust and extensible machine learning infrastructure trained on a variety of data types, to aid in the global response to COVID-19.
We are calling on machine learning practitioners and healthcare professionals who can contribute their expertise to this effort. If you are interested in getting involved in this project by lending your expertise, or sharing data through data-sharing agreements, please reach out to us (contact info is at the bottom of this page); otherwise, feel free to experiment with the code base in this repo. The initial model was built by Blake VanBerlo of the Artificial Intelligence Research and Innovation Lab at the City of London, Canada.
A model has been trained on a dataset composed of X-rays labeled positive for COVID-19 infection, normal X-rays, and X-rays depicting evidence of other pneumonias. Currently, we are using Local Interpretable Model-Agnostic Explanations (i.e. LIME) as the interpretability method being applied to the model. This project is in need of more expertise and more data. This project is in its infancy and not a tool for medical diagnosis. The immediacy of this work cannot be overstated, as any insights derived from this project may be of benefit to healthcare practitioners and researchers as the COVID-19 pandemic continues to evolve.
There have been promising efforts to apply machine learning to aid in the diagnosis of COVID-19 based on CT scans. Despite the success of these methods, the fact remains that COVID-19 is an infection that is likely to be experienced by communities of all sizes. X-rays are inexpensive and quick to perform; therefore, they are more accessible to healthcare providers working in smaller and/or remote regions. Any insights that may be derived as a result of explainability algorithms applied to a successful model will be invaluable to the global effort of identifying and treating cases of COVID-19. This model is a prototype system and not for medical use and does not offer a diagnosis.
- Clone this repository (for help see this tutorial).
- Install the necessary dependencies (listed in
requirements.txt). To do this, open a terminal in
the root directory of the project and run the following:
$ pip install -r requirements.txt - Create a new folder to contain all of your raw data. Set the RAW_DATA field in the PATHS section of config.yml to the address of this new folder.
- Clone the covid-chestxray-dataset repository inside of your RAW_DATA folder. Set the MILA_DATA field in the PATHS section of config.yml to the address of the root directory of the cloned repository (for help see Project Config).
- Clone the Figure1-COVID-chestxray-dataset repository inside of your RAW_DATA folder. Set the FIGURE1_DATA field in the PATHS section of config.yml to the address of the root directory of the cloned repository.
- Download and unzip the RSNA Pneumonia Detection Challenge dataset from Kaggle somewhere on your local machine. Set the RSNA_DATA field in the PATHS section of config.yml to the address of the folder containing the dataset.
- Execute preprocess.py to create Pandas DataFrames of filenames and labels. Preprocessed DataFrames and corresponding images of the dataset will be saved within data/preprocessed/.
- Execute train.py to train the neural network model. The trained model weights will be saved within results/models/, and its filename will resemble the following structure: modelyyyymmdd-hhmmss.h5, where yyyymmdd-hhmmss is the current time. The TensorBoard log files will be saved within results/logs/training/.
- In config.yml, set MODEL_TO_LOAD within PATHS to the path of the model weights file that was generated in step 6 (for help see Project Config). Execute lime_explain.py to generate interpretable explanations for the model's predictions on the test set. See more details in the LIME Section.
- Once you have the appropriate datasets downloaded, execute preprocess.py. See Getting Started for help obtaining and organizing these raw image datasets. If this script ran properly, you should see folders entitled train, test, and val within data/preprocessed. Addionally, you should see 3 files entitled train_set.csv, val_set.csv, and test_set.csv.
- In config.yml, ensure that EXPERIMENT_TYPE within TRAIN is set to 'single_train'.
- Execute train.py. The trained model's weights will be
located in results/models/, and its filename will resemble the
following structure: modelyyyymmdd-hhmmss.h5, where yyyymmdd-hhmmss
is the current time. The model's logs will be located in
results/logs/training/, and its directory name will be the current
time in the same format. These logs contain information about the
experiment, such as metrics throughout the training process on the
training and validation sets, and performance on the test set. The
logs can be visualized by running
TensorBoard locally. See
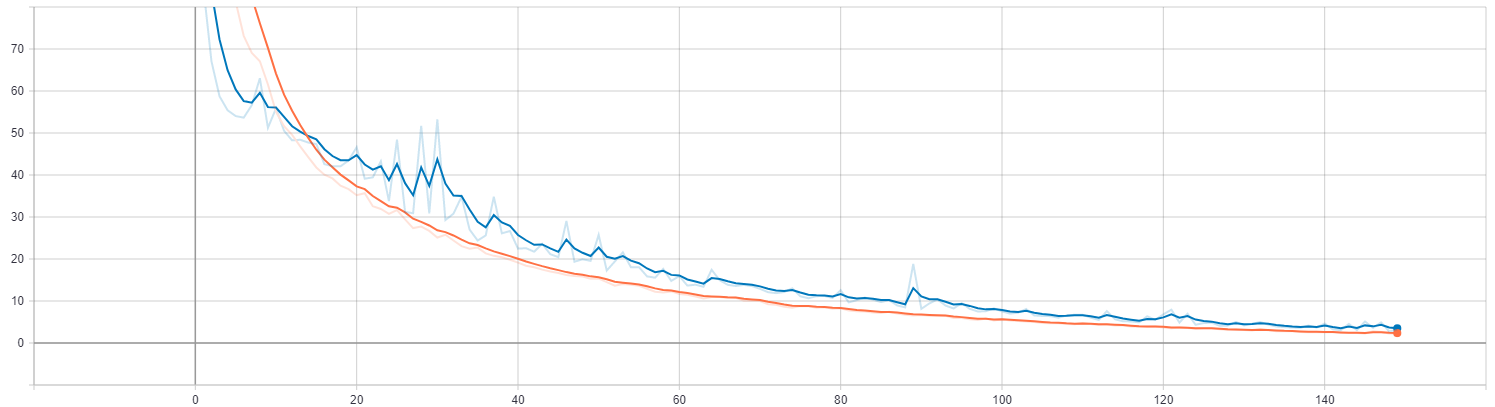
below for an example of a plot from a TensorBoard log file depicting
loss on the training and validation sets versus epoch. Plots
depicting the change in performance metrics throughout the training
process (such as the example below) are available in the SCALARS
tab of TensorBoard.
You can also visualize the trained model's performance on the test set. See below for an example of the ROC Curve and Confusion Matrix based on test set predictions. In our implementation, these plots are available in the IMAGES tab of TensorBoard.
The documentation in this README assumes the user is training a binary classifier, which is set by default in config.yml. The user has the option of training a model to perform binary prediction on whether the X-ray exhibits signs of COVID-19 or training a model to perform multi-class classification to distinguish COVID-19 cases, other pneumonia cases and normal X-rays. For a multi-class model, the output layer is a vector of probabilities outputted by a softmax final layer. In the multi-class scenario, precision, recall and F1-Score are calculated for the COVID-19 class only. To train a multi-class classifier, the user should be aware of the following changes that may be made to config.yml:
- Within the TRAIN section, set the CLASS_MODE field to 'multiclass'. By default, it is set to 'binary'.
- The class names are listed in the CLASSES field of the DATA section.
- The relative weight of classes can be modified by updating the CLASS_MULTIPLIER field in the TRAIN section.
- You can update hyperparameters for the multiclass classification model by setting the fields in the DCNN_MULTICLASS subsection of NN.
- By default, LIME explanations are given for the class of the model's prediction. If you wish to generate LIME explanations for the COVID-19 class only, set COVID_ONLY within LIME to 'true'.
Since the predictions made by this model may be used be healthcare providers to benefit patients, it is imperative that the model's predictions may be explained so as to ensure that the it is making responsible predictions. Model explainability promotes transparency and accountability of decision-making algorithms. Since this model is a neural network, it is difficult to decipher which rules or heuristics it is employing to make its predictions. Since so little is known about presentation of COVID-19, interpretability is all the more important. We used Local Interpretable Model-Agnostic Explanations (i.e. LIME) to explain the predictions of the neural network classifier that we trained. We used the implementation available in the authors' GitHub repository. LIME perturbs the features in an example and fits a linear model to approximate the neural network at the local region in the feature space surrounding the example. It then uses the linear model to determine which features were most contributory to the model's prediction for that example. By applying LIME to our trained model, we can conduct informed feature engineering based on any obviously inconsequential features we see insights from domain experts. For example, we noticed that different characters present on normal X-rays were contributing to predictions based off LIME explanations. To counter this unwanted behaviour, we have taken steps to remove and inpaint textual regions as much as possible. See the steps below to apply LIME to explain the model's predictions on examples in the test set.
- Having previously run preprocess.py and train.py, ensure that data/processed/ contains Test_Set.csv.
- In config.yml, set MODEL_TO_LOAD within PATHS to the path of the model weights file (.h5 file) that you wish to use for prediction.
- Execute lime_explain.py. To
generate explanations for different images in the test set, modify
the following call:
explain_xray(lime_dict, i, save_exp=True). Set i to the index of the test set image you would like to explain and rerun the script. If you are using an interactive console, you may choose to simply call the function again instead of rerunning the script. - Interpret the output of the LIME explainer. An image will have been generated that depicts the superpixels (i.e. image regions) that were most contributory to the model's prediction. Superpixels that contributed toward a prediction of COVID-19 are coloured green and superpixels that contributed against a prediction of COVID-19 are coloured red. The image will be automatically saved in documents/generated_images/, and its filename will resemble the following: original-filename_exp_yyyymmdd-hhmmss.png. See below for examples of this graphic.
It is our hope that healthcare professionals will be able to provide feedback on the model based on their assessment of the quality of these explanations. If the explanations make sense to individuals with extensive experience interpreting X-rays, perhaps certain patterns can be identified as radiological signatures of COVID-19.
Below are examples of LIME explanations. The top two images are explanations of a couple of the binary classifier's predictions. Green regions and red regions identify superpixels that most contributed to and against prediction of COVID-19 respectively. The bottom two images are explanations of a couple of the multi-class classifier's predictions. Green regions and red regions identify superpixels that most contributed to and against the predicted class respectively.
We investigated Grad-CAM as an additional method of explainability. Grad-CAM enables one to visualize the gradient of the label in the final convolutional layer to produce a heatmap depicting regions of the image that were highly important during prediction. The steps to use Grad-CAM in this repository are as follows:
- Follow steps 1 and 2 in LIME Explanations.
- Execute gradcam.py. To generate
explanations for different images in the test set, modify the
following call:
apply_gradcam(lime_dict, i, hm_intensity=0.5, save_exp=True). Set i to the index of the test set image you would like to explain and rerun the script. - Interpret the output of Grad-CAM. Bluer pixels and redder pixels correspond to higher and lower values of the gradient at the final convolutional layer respectively. An image of your heatmap will be saved in documents/generated_images/, and its filename will resemble the following: original-filename_gradcamp_yyyymmdd-hhmmss.png. See below for examples of this graphic.
Not every model trained will perform at the same level on the test set. This procedure enables you to train multiple models and save the one that scored the best result on the test set for a particular metric that you care about optimizing.
- Follow step 1 in Train a model and visualize results.
- In config.yml, set EXPERIMENT_TYPE within TRAIN to 'multi_train'.
- Decide which metrics you would like to optimize and in what order. In config.yml, set METRIC_PREFERENCE within TRAIN to your chosen metrics, in order from most to least important. For example, if you decide to select the model with the best AUC on the test set, set the first element in this field to 'AUC'.
- Decide how many models you wish to train. In config.yml, set NUM_RUNS within TRAIN to your chosen number of training sessions. For example, if you wish to train 10 models, set this field to 10.
- Execute train.py. The weights of the model that had the best performance on the test set for the metric you specified will be located in results/models/training/, and its filename will resemble the following structure: modelyyyymmdd-hhmmss.h5, where yyyymmdd-hhmmss is the current time. Logs for each run will be located in results/logs/training/, and their names will be the current time in the same format. The log directory that has the same name as the model weights file corresponds to the run with the best metrics.
Hyperparameter tuning is an important part of the standard machine learning workflow. Our implementation allows users to conduct a random hyperparameter search. Many fields in the TRAIN and NN sections of config.yml are considered to be hyperparameters. With the help of the HParam Dashboard, one can see the effect of different combinations of hyperparameters on the model's test set performance metrics.
In our random hyperparameter search, we study the effects of x random combinations of hyperparameters by training the model y times for each of the x combinations and recording the results. See the steps below on how to conduct a random hyperparameter search with our implementation for the following 4 hyperparameters: dropout, learning rate, conv blocks, and optimizer.
- In the in the HP_SEARCH section of config.yml, set
the number of random combinations of hyperparameters you wish to
study and the number of times you would like to train the model for
each combination.
COMBINATIONS: 50 REPEATS: 2 - Set the ranges of hyperparameters you wish to study in the RANGES
subsection of the HP_SEARCH section of config.yml.
Consider whether your hyperparameter ranges are continuous (i.e.
real), discrete or integer ranges and whether any need to be
investigated on the logarithmic scale.
DROPOUT: [0.2, 0.5] # Real range LR: [-4.0, -2.5] # Real range on logarithmic scale (10^x) CONV_BLOCKS: [2, 4] # Integer range OPTIMIZER: ['adam', 'sgd'] # Discrete range - Within the random_hparam_search() function defined in
train.py, add your hyperparameters as HParam objects
to the list of hyperparameters being considered (if they are not
already there).
HPARAMS.append(hp.HParam('DROPOUT', hp.RealInterval(hp_ranges['DROPOUT'][0], hp_ranges['DROPOUT'][1]))) HPARAMS.append(hp.HParam('LR', hp.RealInterval(hp_ranges['LR'][0], hp_ranges['LR'][1]))) HPARAMS.append(hp.HParam('CONV_BLOCKS', hp.IntInterval(hp_ranges['CONV_BLOCKS'][0], hp_ranges['CONV_BLOCKS'][1]))) HPARAMS.append(hp.HParam('OPTIMIZER', hp.Discrete(hp_ranges['OPTIMIZER']))) - In the appropriate location (varies by hyperparameter), ensure that
you assign the hyperparameters based on the random combination. If
the hyperparameter already exists in the NN section of
config.yml, this will be done for you automatically. In
our example, all of these hyperparameters are already in the NN
section of the project config, so it will be done automatically. If the hyperparameter is not in the NN section, assign the
hyperparameter value within the code for a single training run in the
random_hparam_search() method in train.py (for
example, batch size).
cfg['TRAIN']['BATCH_SIZE'] = hparams['BATCH_SIZE'] - In config.yml, set EXPERIMENT_TYPE within the TRAIN section to 'hparam_search'.
- Execute train.py. The experiment's logs will be located in results/logs/hparam_search/, and the directory name will be the current time in the following format: yyyymmdd-hhmmss. These logs contain information on test set metrics with models trained on different combinations of hyperparameters. The logs can be visualized by running TensorBoard locally and clicking on the HPARAM tab.
Once a trained model is produced, the user may wish to obtain predictions and explanations for a list of images. The steps below detail how to run prediction for all images in a specified folder, given a trained model and serialized LIME Image Explainer object.
- Ensure that you have already run lime_explain.py after training your model, as it will have generated and saved a LIME Image Explainer object at data/interpretability/lime_explainer.pkl.
- Specify the path of the image folder containing the X-ray images you wish to predict and explain. In config.yml, set BATCH_PRED_IMGS within PATHS to the path of your image folder.
- In config.yml, set MODEL_TO_LOAD within PATHS to the path of the model weights file (.h5 file) that you wish to use for prediction.
- Execute predict.py. Running this script will preprocess images, run prediction for all images, and run LIME to explain the predictions. Prediction results will be saved in a .csv file, which will be located in results/predictions/yyyymmdd-hhmmss (where yyyymmdd-hhmmss is the time at which the script was executed), and will be called predictions.csv. The .csv file will contain image file names, predicted class, model output probabilities, and the file name of the corresponding explanation. The images depicting LIME explanations will be saved in the same folder.
We ran several of our training experiments on Azure cloud compute instances. To do this, we created Jupyter notebooks to define and run experiments in Azure, and Python scripts corresponding to pipeline steps. We included these files in the azure/ folder, in case they may benefit any parties hoping to extend our project. Note that Azure is not required to run this project - all Python files necessary to get started are in the src/ folder. If you plan on using the Azure machine learning pipelines defined in the azure/ folder, be sure to install the azureml-sdk and azureml_widgets packages.
The project looks similar to the directory structure below. Disregard any .gitkeep files, as their only purpose is to force Git to track empty directories. Disregard any ._init_.py files, as they are empty files that enable Python to recognize certain directories as packages.
├── azure <- folder containing Azure ML pipelines
├── data
│ ├── interpretability <- Interpretability files
│ └── processed <- Products of preprocessing
|
├── documents
| ├── generated_images <- Visualizations of model performance, experiments
| └── readme_images <- Image assets for README.md
├── results
│ ├── logs <- TensorBoard logs
│ └── models <- Trained model weights
|
├── src
│ ├── custom <- Custom TensorFlow components
| | └── metrics.py <- Definition of custom TensorFlow metrics
│ ├── data <- Data processing
| | └── preprocess.py <- Main preprocessing script
│ ├── interpretability <- Model interpretability scripts
| | ├── gradcam.py <- Script for generating Grad-CAM explanations
| | └── lime_explain.py <- Script for generating LIME explanations
│ ├── models <- TensorFlow model definitions
| | └── models.py <- Script containing model definition
| ├── visualization <- Visualization scripts
| | └── visualize.py <- Script for visualizing model performance metrics
| ├── predict.py <- Script for running batch predictions
| └── train.py <- Script for training model on preprocessed data
|
├── .gitignore <- Files to be be ignored by git.
├── config.yml <- Values of several constants used throughout project
├── LICENSE <- Project license
├── README.md <- Project description
└── requirements.txt <- Lists all dependencies and their respective versions
Many of the components of this project are ready for use on your X-ray datasets. However, this project contains several configurable variables that are defined in the project config file: config.yml. When loaded into Python scripts, the contents of this file become a dictionary through which the developer can easily access its members.
For user convenience, the config file is organized into major steps in our model development pipeline. Many fields need not be modified by the typical user, but others may be modified to suit the user's specific goals. A summary of the major configurable elements in this file is below.
- RAW_DATA: Path to parent folder containing all downloaded datasets (i.e. MILA_DATA, FIGURE1_DATA, RSNA_DATA)
- MILA_DATA: Path to folder containing Mila COVID-19 image dataset
- FIGURE1_DATA: Path to folder containing Figure 1 image dataset
- RSNA_DATA: Path to folder containing RSNA Pneumonia Detection Challenge dataset
- MODEL_WEIGHTS: Path at which to save trained model's weights
- MODEL_TO_LOAD: Path to the trained model's weights that you would like to load for prediction
- BATCH_PRED_IMGS: Path to folder containing images for batch prediction
- BATCH_PREDS: Path to folder for outputting batch predictions and explanations
- IMG_DIM: Desired target size of image after preprocessing
- VIEWS: List of types of chest X-ray views to include. By default, posteroanterior and anteroposterior views are included.
- VAL_SPLIT: Fraction of the data allocated to the validation set
- TEST_SPLIT: Fraction of the data allocated to the test set
- NUM_RSNA_IMGS: Number of images from the RSNA dataset that you wish to include in your assembled dataset
- CLASSES: This is an ordered list of class names. Must be the same length as the number of classes you wish to distinguish.
- CLASS_MODE: The type of classification to be performed. Should be set before performing preprocessing. Set to either 'binary' or 'multiclass'.
- CLASS_MULTIPLIER: A list of coefficients to multiply the computed class weights by during computation of loss function. Must be the same length as the number of classes.
- EXPERIMENT_TYPE: The type of training experiment you would like to perform if executing train.py. For now, the only choice is 'single_train'.
- BATCH_SIZE: Mini-batch size during training
- EPOCHS: Number of epochs to train the model for
- THRESHOLDS: A single float or list of floats in range [0, 1] defining the classification threshold. Affects precision and recall metrics.
- PATIENCE: Number of epochs to wait before freezing the model if validation loss does not decrease.
- IMB_STRATEGY: Class imbalancing strategy to employ. In our dataset, the ratio of positive to negative ground truth was very low, prompting the use of these strategies. Set either to 'class_weight' or 'random_oversample'.
- METRIC_PREFERENCE: A list of metrics in order of importance (from left to right) to guide selection of the best model during a 'multi_train' experiment
- NUM_RUNS: The number of times to train a model in the 'multi_train' experiment
- NUM_GPUS: The number of GPUs to distribute training over
- DCNN_BINARY: Contains definitions of configurable hyperparameters
associated with a custom deep convolutional neural network for binary
classification. The values currently in this section were the optimal
values for our dataset informed by heuristically selecting
hyperparameters.
- KERNEL_SIZE: Kernel size for convolutional layers
- STRIDES: Size of strides for convolutional layers
- INIT_FILTERS: Number of filters for first convolutional layer
- FILTER_EXP_BASE: Base of exponent that determines number of filters in successive convolutional layers. For layer i, # filters = INIT_FILTERS * (FILTER_EXP_BASE) i
- CONV_BLOCKS: The number of convolutional blocks. Each block contains a 2D convolutional layer, a batch normalization layer, activation layer, and a maxpool layer.
- NODES_DENSE0: The number of nodes in the fully connected layer following flattening of parameters
- LR: Learning rate
- OPTIMIZER: Optimization algorithm
- DROPOUT: Dropout rate
- L2_LAMBDA: L2 regularization parameter
- DCNN_MULTICLASS: Contains definitions of configurable hyperparameters associated with a custom deep convolutional neural network for multi-class classification. The fields are identical to those in the DCNN_BINARY subsection.
- KERNEL_WIDTH: Affects size of neighbourhood around which LIME samples for a particular example. In our experience, setting this within the continuous range of [1.5, 2.0] is large enough to produce stable explanations, but small enough to avoid producing explanations that approach a global surrogate model.
- FEATURE_SELECTION: The strategy to select features for LIME explanations. Read the LIME creators' documentation for more information.
- NUM_FEATURES: The number of features to include in a LIME explanation
- NUM_SAMPLES: The number of samples used to fit a linear model when explaining a prediction using LIME COVID_ONLY: Set to 'true' if you want explanations to be provided for the predicted logit corresponding to the "COVID-19" class, despite the model's prediction. If set to 'false', explanations will be provided for the logit corresponding to the predicted class.
- METRICS: List of metrics on validation set to monitor in hyperparameter search. Can be any combination of {'accuracy', 'loss', 'recall', 'precision', 'auc'}
- COMBINATIONS: Number of random combinations of hyperparameters to try in hyperparameter search
- REPEATS: Number of times to repeat training per combination of hyperparameters
- RANGES: Ranges defining possible values that hyperparameters may take. Be sure to check train.py to ensure that your ranges are defined correctly as real or discrete intervals (see Random Hyperparameter Search for an example).
- THRESHOLD: Classification threshold for prediction
Matt Ross
Manager, Artificial Intelligence
Information Technology Services
City Manager’s Office
The Corporation of the City of London
201 Queens Ave. Suite 300, London, ON. N6A 1J1
C: 226.448.9113 | maross@london.ca
Blake VanBerlo
Data Scientist
City of London Research and Innovation Lab
C: vanberloblake@gmail.com