DrugGEN: Target Centric De Novo Design of Drug Candidate Molecules with Graph Generative Deep Adversarial Networks
Please see our most up-to-date document (pre-print) from 15.02.2023 here: 2302.07868.pdf, arXiv link
Discovering novel drug candidate molecules is one of the most fundamental and critical steps in drug development. Generative deep learning models, which create synthetic data given a probability distribution, have been developed with the purpose of picking completely new samples from a partially known space. Generative models offer high potential for designing de novo molecules; however, in order for them to be useful in real-life drug development pipelines, these models should be able to design target-specific molecules, which is the next step in this field. In this study, we propose DrugGEN, for the de novo design of drug candidate molecules that interact with selected target proteins. The proposed system represents compounds and protein structures as graphs and processes them via serially connected two generative adversarial networks comprising graph transformers. DrugGEN is trained using a large dataset of compounds from ChEMBL and target-specific bioactive molecules, to design effective and specific inhibitory molecules against the AKT1 protein, which has critical importance for developing treatments against various types of cancer. On fundamental benchmarks, DrugGEN models have either competitive or better performance against other methods. To assess the target-specific generation performance, we conducted further in silico analysis with molecular docking and deep learning-based bioactivity prediction. Results indicate that de novo molecules have high potential for interacting with the AKT1 protein structure in the level of its native ligand. DrugGEN can be used to design completely novel and effective target-specific drug candidate molecules for any druggable protein, given target features and a dataset of experimental bioactivities. Code base, datasets, results and trained models of DrugGEN are available in this repository.
Our up-to-date pre-print is shared here
Fig. 1. (A) Generator (G1) of the GAN1 consists of an MLP and graph transformer encoder module. The generator encodes the given input into a new representation; (B) the MLP-based discriminator (D1) of GAN1 compares the generated de novo molecules to the real ones in the training dataset, scoring them for their assignment to the classes of “real” and “fake” molecules; (C) Generator (G2) of GAN2 makes use of the transformer decoder architecture to process target protein features and GAN1 generated de novo molecules together. The output of the generator two (G2) is the modified molecules, based on the given protein features; (D) the second discriminator (D2) takes the modified de novo molecules and known inhibitors of the given target protein and scores them for their assignment to the classes of “real” and “fake” inhibitors.
Given a random noise z, the first generator G1 (below, on the left side) creates annotation and adjacency matrices of a supposed molecule. G1 processes the input by passing it through a multi-layer perceptron (MLP). The input is then fed to the transformer encoder module Vaswani et al., (2017), which has a depth of 8 encoder layers with 8 multi-head attention heads for each. In the graph transformer setting, Q, K and V are the variables representing the annotation matrix of the molecule. After the final products are created in the attention mechanism, both the annotation and adjacency matrices are forwarded to layer normalization and then summed with the initial matrices to create a residual connection. These matrices are fed to separate feedforward layers, and finally, given to the discriminator network D1 together with real molecules.
The second generator G2 (below, on the right side) modifies molecules that were previously generated by G1, with the aim of generating binders for the given target protein. G2 module utilizes the transformer decoder architecture. This module has a depth of 8 decoder layers and uses 8 multi-head attention heads for each. G2 takes both G1(z), which is data generated by G1, and the protein features as input. Interactions between molecules and proteins are processed inside the multi-head attention module via taking their scaled dot product, and thus, new molecular graphs are created. Apart from the attention mechanism, further processing of the molecular matrices follows the same workflow as the transformer encoder. The output of this module are the final product of the DrugGEN model and are forwarded to D2.
| First Generator | Second Generator |
|---|---|
 |
 |
- DrugGEN-Prot (the default model) is composed of two GANs. It incorporates protein features to the transformer decoder module of GAN2 (together with the de novo molecules generated by GAN1) to direct the target centric molecule design. The information provided above belongs to this model.
- DrugGEN-CrossLoss is composed of only one GAN. The input of the GAN1 generator is the real molecules (ChEMBL) dataset (to ease the learning process) and the GAN1 discriminator compares the generated molecules with the real inhibitors of the given target protein.
- DrugGEN-Ligand is composed of two GANs. It incorporates AKT1 inhibitor molecule features as the input of the GAN2-generator’s transformer decoder instead of the protein features in the default model.
- DrugGEN-RL utilizes the same architecture as the DrugGEN-Ligand model. It uses reinforcement learning (RL) to avoid using molecular scaffolds that are already presented in the training set.
- DrugGEN-NoTarget is composed of only one GAN. This model only focuses on learning the chemical properties from the ChEMBL training dataset, as a result, there is no target-specific generation.
We provide the implementation of the DrugGEN, along with scripts from PyTorch Geometric framework to generate and run. The repository is organised as follows:
data contains:
- Raw dataset files, which should be text files containing SMILES strings only. Raw datasets preferably should not contain stereoisomeric SMILES to prevent Hydrogen atoms to be included in the final graph data.
- Constructed graph datasets (.pt) will be saved in this folder along with atom and bond encoder/decoder files (.pk).
experiments contains:
logsfolder. Model loss and performance metrics will be saved in this directory in seperate files for each model.tboard_outputfolder. Tensorboard files will be saved here if TensorBoard is used.modelsfolder. Models will be saved in this directory at last or preferred steps.samplesfolder. Molecule samples will be saved in this folder.inferencefolder. Molecules generated in inference mode will be saved in this folder.
Python scripts:
layers.pycontains transformer encoder and transformer decoder implementations.main.pycontains arguments and this file is used to run the model.models.pyhas the implementation of the Generators and Discriminators which are used in GAN1 and GAN2.new_dataloader.pyconstructs the graph dataset from given raw data. Uses PyG based data classes.trainer.pyis the training and testing file for the model. Workflow is constructed in this file.utils.pycontains performance metrics from several other papers and some unique implementations. (De Cao et al, 2018; Polykovskiy et al., 2020)
Three different data types (i.e., compound, protein, and bioactivity) were retrieved from various data sources to train our deep generative models. GAN1 module requires only compound data while GAN2 requires all of three data types including compound, protein, and bioactivity.
- Compound data includes atomic, physicochemical, and structural properties of real drug and drug candidate molecules. ChEMBL v29 compound dataset was used for the GAN1 module. It consists of 1,588,865 stable organic molecules with a maximum of 45 atoms and containing C, O, N, F, Ca, K, Br, B, S, P, Cl, and As heavy atoms.
- Protein data was retrieved from Protein Data Bank (PDB) in biological assembly format, and the coordinates of protein-ligand complexes were used to construct the binding sites of proteins from the bioassembly data. The atoms of protein residues within a maximum distance of 9 A from all ligand atoms were recorded as binding sites. GAN2 was trained for generating compounds specific to the target protein AKT1, which is a member of serine/threonine-protein kinases and involved in many cancer-associated cellular processes including metabolism, proliferation, cell survival, growth and angiogenesis. Binding site of human AKT1 protein was generated from the kinase domain (PDB: 4GV1).
- Bioactivity data of AKT target protein was retrieved from large-scale ChEMBL bioactivity database. It contains ligand interactions of human AKT1 (CHEMBL4282) protein with a pChEMBL value equal to or greater than 6 (IC50 <= 1 µM) as well as SMILES information of these ligands. The dataset was extended by including drug molecules from DrugBank database known to interact with human AKT proteins. Thus, a total of 1,600 bioactivity data points were obtained for training the AKT-specific generative model.
More details on the construction of datasets can be found in our paper referenced above.
DrugGEN has been implemented and tested on Ubuntu 18.04 with python >= 3.9. It supports both GPU and CPU inference.
Clone the repo:
git clone https://github.com/HUBioDataLab/DrugGEN.git
You can set up the environment using either conda or pip.
Here is with conda:
# set up the environment (installs the requirements):
conda env create -f DrugGEN/dependencies.yml
# activate the environment:
conda activate druggenHere is with pip using virtual environment:
python -m venv DrugGEN/.venv
./Druggen/.venv/bin/activate
pip install -r DrugGEN/requirements.txt# Download input files:
cd DrugGEN/data
bash dataset_download.sh
cd
# DrugGEN can be trained with the one-liner:
python DrugGEN/main.py --submodel="CrossLoss" --mode="train" --raw_file="DrugGEN/data/chembl_train.smi" --dataset_file="chembl45_train.pt" --drug_raw_file="DrugGEN/data/akt_train.smi" --drug_dataset_file="drugs_train.pt" --max_atom=45
** Explanations of arguments can be found below:
Model arguments:
--submodel SUBMODEL Choose the submodel for training
--act ACT Activation function for the model
--z_dim Z_DIM Prior noise for the first GAN
--max_atom MAX ATOM Maximum atom number for molecules must be specified
--lambda_gp LAMBDA_GP Gradient penalty lambda multiplier for the first GAN
--dim DIM Dimension of the Transformer models for both GANs
--depth DEPTH Depth of the Transformer model from the first GAN
--heads HEADS Number of heads for the MultiHeadAttention module from the first GAN
--dec_depth DEC_DEPTH Depth of the Transformer model from the second GAN
--dec_heads DEC_HEADS Number of heads for the MultiHeadAttention module from the second GAN
--mlp_ratio MLP_RATIO MLP ratio for the Transformers
--dis_select DIS_SELECT Select the discriminator for the first and second GAN
--init_type INIT_TYPE Initialization type for the model
--dropout DROPOUT Dropout rate for the encoder
--dec_dropout DEC_DROPOUT Dropout rate for the decoder
Training arguments:
--batch_size BATCH_SIZE Batch size for the training
--epoch EPOCH Epoch number for Training
--warm_up_steps Warm up steps for the first GAN
--g_lr G_LR Learning rate for G
--g2_lr G2_LR Learning rate for G2
--d_lr D_LR Learning rate for D
--d2_lr D2_LR Learning rate for D2
--n_critic N_CRITIC Number of D updates per each G update
--beta1 BETA1 Beta1 for Adam optimizer
--beta2 BETA2 Beta2 for Adam optimizer
--clipping_value Clipping value for the gradient clipping process
--resume_iters Resume training from this step for fine tuning if desired
Dataset arguments:
--features FEATURES Additional node features (Boolean) (Please check new_dataloader.py Line 102)
- First, download the weights of the chosen trained model from trained models, and place it in the folder: "DrugGEN/experiments/models/".
- After that, please run the code below:
python DrugGEN/main.py --submodel="{Chosen model name}" --mode="inference" --inference_model="DrugGEN/experiments/models/{Chosen model name}"- SMILES representation of the generated molecules will be saved into the file: "DrugGEN/experiments/inference/{Chosen model name}/denovo_molecules.txt".
- SMILES notations of 50,000 de novo generated molecules from DrugGEN models (10,000 from each) can be downloaded from here.
- We first filtered the 50,000 de novo generated molecules by applying Lipinski, Veber and PAINS filters; and 43,000 of them remained in our dataset after this operation (SMILES notations of filtered de novo molecules).
- We run our deep learning-based drug/compound-target protein interaction prediction system DEEPScreen on 43,000 filtered molecules. DEEPScreen predicted 18,000 of them as active against AKT1, 301 of which received high confidence scores (> 80%) (SMILES notations of DeepScreen predicted actives).
- At the same time, we performed a molecular docking analysis on these 43,000 filtered de novo molecules against the crystal structure of AKT1, and found that 118 of them had sufficiently low binding free energies (< -9 kcal/mol) (SMILES notations of de novo molecules with low binding free energies).
- Finally, de novo molecules to effectively target AKT1 protein are selected via expert curation from the dataset of molecules with binding free energies lower than -9 kcal/mol. The structural representations of the selected molecules are shown in the figure below (SMILES notations of the expert selected de novo AKT1 inhibitor molecules).
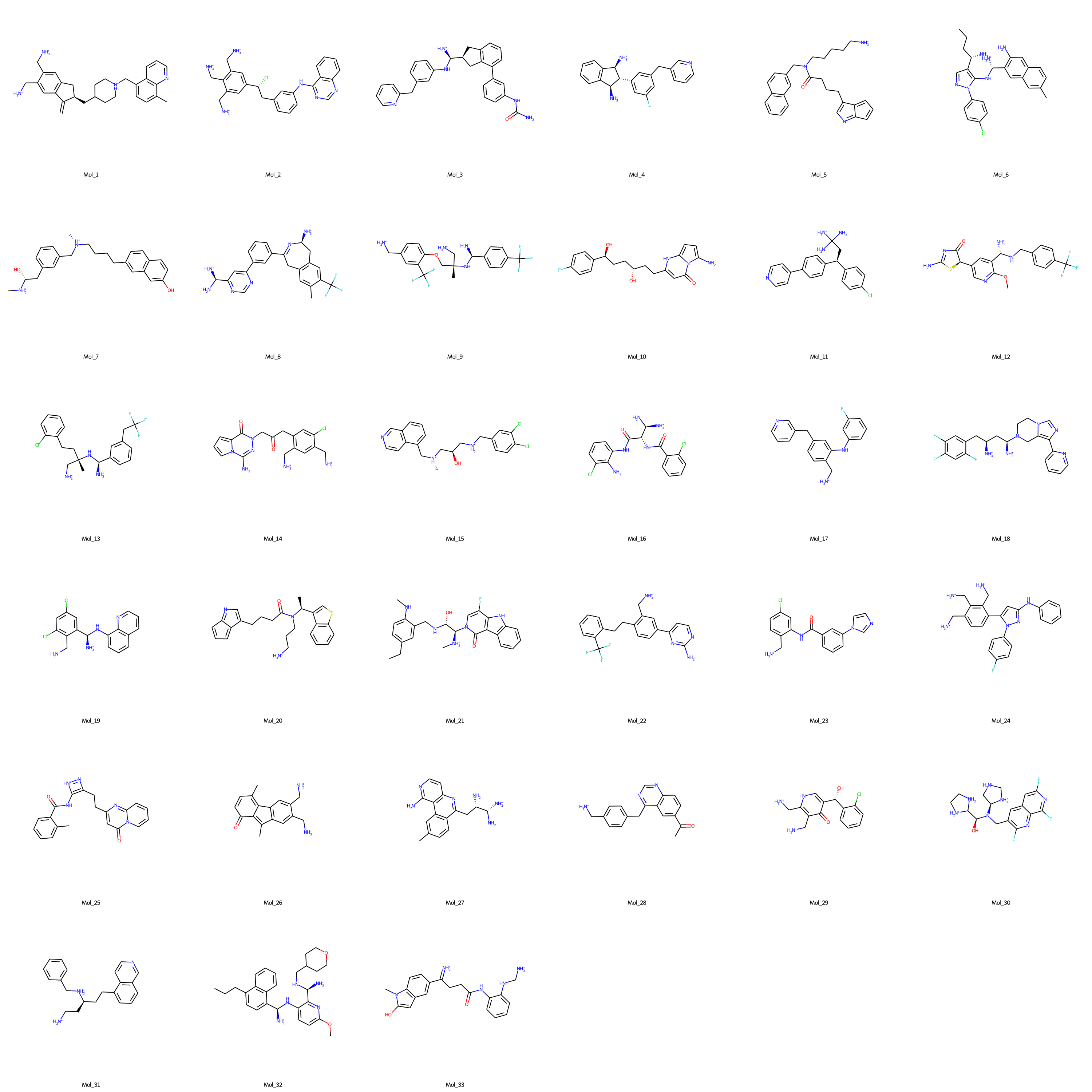 Fig. 2. Promising de novo molecules to effectively target AKT1 protein (generated by DrugGEN models), selected via expert curation from the dataset of molecules with sufficiently low binding free energies (< -9 kcal/mol) in the molecular docking experiment.
Fig. 2. Promising de novo molecules to effectively target AKT1 protein (generated by DrugGEN models), selected via expert curation from the dataset of molecules with sufficiently low binding free energies (< -9 kcal/mol) in the molecular docking experiment.
- 15/02/2023: Our pre-print is shared here.
- 01/01/2023: Five different DrugGEN models are released.
@misc{nl2023target,
doi = {10.48550/ARXIV.2302.07868},
title={Target Specific De Novo Design of Drug Candidate Molecules with Graph Transformer-based Generative Adversarial Networks},
author={Atabey Ünlü and Elif Çevrim and Ahmet Sarıgün and Hayriye Çelikbilek and Heval Ataş Güvenilir and Altay Koyaş and Deniz Cansen Kahraman and Abdurrahman Olğaç and Ahmet Rifaioğlu and Tunca Doğan},
year={2023},
eprint={2302.07868},
archivePrefix={arXiv},
primaryClass={cs.LG}
}Ünlü, A., Çevrim, E., Sarıgün, A., Çelikbilek, H., Güvenilir, H.A., Koyaş, A., Kahraman, D.C., Olğaç, A., Rifaioğlu, A., Doğan, T. (2023). Target Specific De Novo Design of Drug Candidate Molecules with Graph Transformer-based Generative Adversarial Networks. arXiv preprint arXiv:2302.07868.
In each file, we indicate whether a function or script is imported from another source. Here are some excellent sources from which we benefit from:
- Molecule generation GAN schematic was inspired from MolGAN.
- MOSES was used for performance calculation (MOSES Script are directly embedded to our code due to current installation issues related to the MOSES repo).
- PyG was used to construct the custom dataset.
- Transformer architecture was taken from Vaswani et al. (2017).
- Graph Transformer Encoder architecture was taken from Dwivedi & Bresson (2021) and Vignac et al. (2022) and modified.
Our initial project repository was this one.
Copyright (C) 2023 HUBioDataLab
This program is free software: you can redistribute it and/or modify it under the terms of the GNU General Public License as published by the Free Software Foundation, either version 3 of the License, or (at your option) any later version.
This program is distributed in the hope that it will be useful, but WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the GNU General Public License for more details.
You should have received a copy of the GNU General Public License along with this program. If not, see http://www.gnu.org/licenses/.


