The goal of adas.utils is to provide some utility functions to be used in the course Analysis of Data and Statistics at the University of Trento, Italy. Course contents are on https://paolobosetti.quarto.pub/ADAS/.
You can install the development version of adas.utils from GitHub with:
# install.packages("devtools")
devtools::install_github("pbosetti/adas.utils", build_vignettes = TRUE)On https://paolobosetti.quarto.pub/data.html there are a list of
example datasets to be used during the course. You can load them with
the examples_url function:
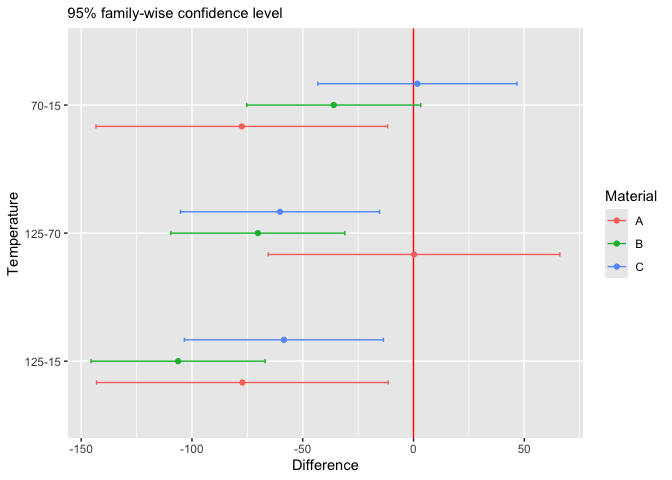
examples_url("battery.dat") |> read.table(header=T) |> head()
#> RunOrder StandardOrder Temperature Material Repeat Response
#> 1 34 1 15 1 1 130
#> 2 25 2 70 1 1 34
#> 3 16 3 125 1 1 20
#> 4 7 4 15 2 1 150
#> 5 8 5 70 2 1 136
#> 6 1 6 125 2 1 25The Chauvenet’s criterion is a method to identify possible outliers in a sample. Here is an example:
x <- rnorm(100)
x[50] <- 10
chauvenet(x)
#> Chauvenet's criterion for sample x
#> Suspect outlier: 50, value 10
#> Expected frequency: 5.61978810813674e-11, threshold: 0.5
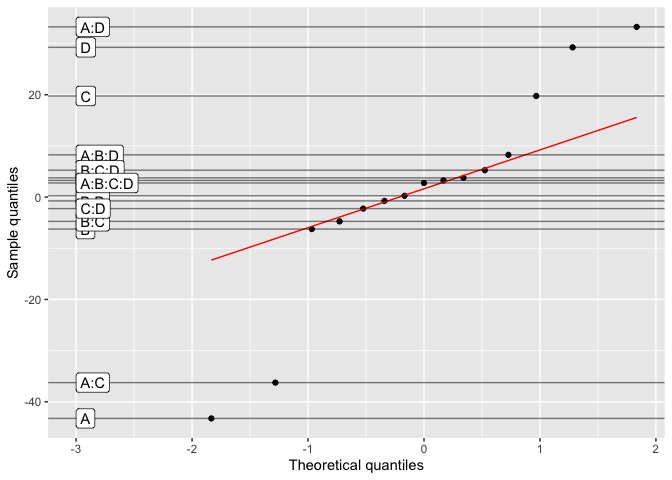
#> Decision: reject itDaniel’s plot is a QQ plot of the effects of a non-replicated factorial model. Here is an example:
daniel_plot_qq(lm(Y~A*B*C*D, data=filtration))The pareto_chart function is a generic function that creates a Pareto
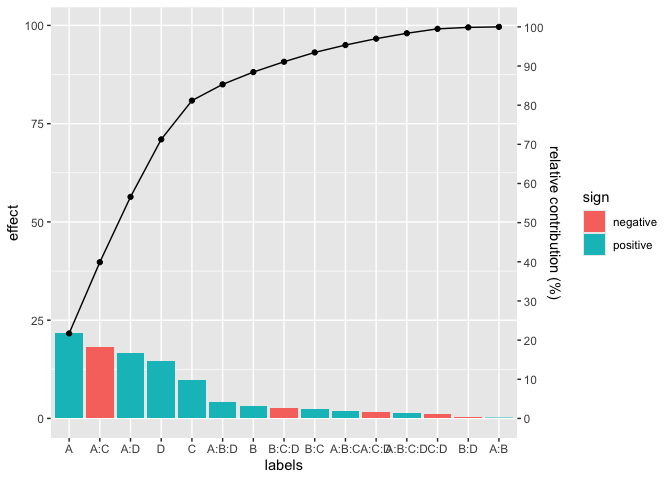
chart either from a general data frame or from the effects of a lm
object. Here is an example:
library(tidyverse)
#> ── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ──
#> ✔ dplyr 1.1.4 ✔ readr 2.1.5
#> ✔ forcats 1.0.0 ✔ stringr 1.5.1
#> ✔ ggplot2 3.5.1 ✔ tibble 3.2.1
#> ✔ lubridate 1.9.3 ✔ tidyr 1.3.1
#> ✔ purrr 1.0.2
#> ── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
#> ✖ tidyr::extract() masks magrittr::extract()
#> ✖ dplyr::filter() masks stats::filter()
#> ✖ dplyr::lag() masks stats::lag()
#> ✖ purrr::set_names() masks magrittr::set_names()
#> ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errors
set.seed(1)
tibble(
val=rnorm(10, sd=5),
cat=LETTERS[1:length(val)]
) %>%
pareto_chart(labels=cat, values=val)# For a linear model:
pareto_chart(lm(Y~A*B*C*D, data=filtration))There is a host of functions to be used in the context of Design of
Experiments. Here is an example to prepare a design matrix for a
fp_design_matrix(5) %>%
fp_fraction(~A*B*C*D) %>%
fp_fraction(~B*C*D*E)
#> Factorial Plan Design Matrix
#> Defining Relationship: ~ A * B * C * D * E
#> Factors: A B C D E
#> Levels: -1 1
#> Fraction: I=ABCD I=BCDE
#> Type: plain
#>
#> # A tibble: 8 × 12
#> StdOrder RunOrder .treat .rep A B C D E Y ABCD BCDE
#> <int> <int> <chr> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <lgl> <dbl> <dbl>
#> 1 1 9 (1) 1 -1 -1 -1 -1 -1 NA 1 1
#> 2 7 14 bc 1 -1 1 1 -1 -1 NA 1 1
#> 3 11 10 bd 1 -1 1 -1 1 -1 NA 1 1
#> 4 13 31 cd 1 -1 -1 1 1 -1 NA 1 1
#> 5 20 4 abe 1 1 1 -1 -1 1 NA 1 1
#> 6 22 13 ace 1 1 -1 1 -1 1 NA 1 1
#> 7 26 26 ade 1 1 -1 -1 1 1 NA 1 1
#> 8 32 24 abcde 1 1 1 1 1 1 NA 1 1You can also prepare a design matrix for a
fp_design_matrix(3, rep=2) %>%
fp_augment_center(rep=5) %>%
fp_augment_axial(rep=2)
#> Factorial Plan Design Matrix
#> Defining Relationship: ~ A * B * C
#> Factors: A B C
#> Levels: -1 1
#> Fraction: NA
#> Type: composite
#>
#> # A tibble: 45 × 8
#> StdOrder RunOrder .treat .rep A B C Y
#> <int> <int> <chr> <int> <dbl> <dbl> <dbl> <lgl>
#> 1 1 10 (1) 1 -1 -1 -1 NA
#> 2 2 7 a 1 1 -1 -1 NA
#> 3 3 3 b 1 -1 1 -1 NA
#> 4 4 16 ab 1 1 1 -1 NA
#> 5 5 6 c 1 -1 -1 1 NA
#> 6 6 8 ac 1 1 -1 1 NA
#> 7 7 2 bc 1 -1 1 1 NA
#> 8 8 13 abc 1 1 1 1 NA
#> 9 9 12 (1) 2 -1 -1 -1 NA
#> 10 10 11 a 2 1 -1 -1 NA
#> # ℹ 35 more rowsPaolo Bosetti, University of Trento