#RAT - The Recombination Analysis Tool
Bioinformatics RAT paper (Etherington et al., 2005) can be found here
The Recombination Analysis Tool (RAT) is an uncomplicated and intuitive cross-platform Java-based application intended for high-throughput, distance-based analysis of both DNA and protein multiple sequence alignments, in any one of seven different file formats. All of RAT's operations are carried out through GUI's, and all output can be saved as data files (.txt, .xls, .csv), or .jpg files in the case of graphical outputs. RAT is intuitive and easy to use, and only requires a minimum of input from the user. All the parameters have default values, all of which may be changed by the user.
You can clone this repository and build the source code yourself (no external libraries neede), or you can use the RAT.jar file found in the 'dist' directory.
RAT comes packaged as a .jar file, and is tested on Java 1.6 and Java 1.7.
Move Rat.jar to a suitable location and double-click the jar file to start. If you are presented with an 'Open with...' box, find the location of javaw.exe (often found in the system32 folder on Windows machines) and use that. Alternatively, you can start RAT from the command line using: java -jar RAT.jar
##USER GUIDE
You will be presented with the main RAT Graphic User Interface (GUI) as follows:
** Figure 1. The main RAT GUI.**
Click on the "Choose Alignment File" and pick a DNA or protein multiple alignment file. RAT handles most of the popular alignment formats (.fas, .phy, .gde (% and # versions), etc). Due to their simple format, fastA files load the quickest.
The RAT GUI should now look something similar to this:
Figure 2. The RAT GUI with a loaded file and default parameters shown.
The name of your file should be below the file chooser button, all the sequences in the alignment should be listed in the drop-down menu below 'Test sequence name:', there should be default parameters in the 'Window size', 'Increment size', 'Start at position', and 'End at position' boxes.
The number in the 'Start at position' should be 1 and the number in the 'End at position' should be the length of the sequences in the alignment file. The 'Window size' should be 10% of the sequence length and the 'Increment size' should be half of 'Window size'.
There should also be default parameters in the 'Auto Search' fields below (see Auto search below for a discussion of these).
SINGLE-SEQUENCE VIEWER
If you have a sequence that you are particularly interested in examining for recombination, then you can user the Single-sequence Viewer option. From the 'Test sequence name:' drop-down menu, select your sequence and click 'Execute'. After a few seconds (depending on the size of your alignment file), you should see the Sequence Viewer, like so:
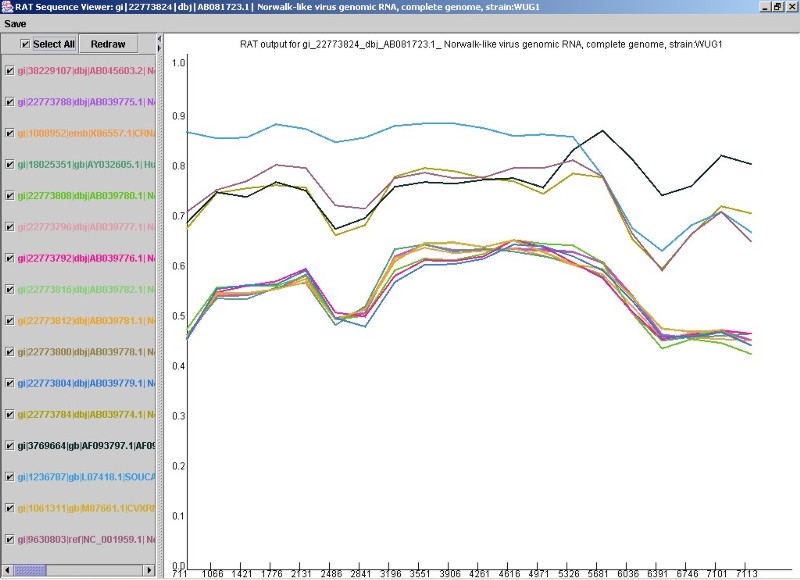
Figure 3. The RAT sequence viewer.
The coloured lines on the graph (Fig. 3) represent the genetic distance (Y-axis) of each sequence in the alignment file to that of the sequence you chose as your 'Test sequence' earlier. The X-axis represents the location on the sequence. Note that each sequence name is checked in the sequence list pane on the right. You can uncheck sequences that you are not interested in and click on 'Redraw' to remove those sequences from the graph pane on the right. The colour of the line on the graph is the same as the colour of its sequence name on the left. You can also add sequences back on to the graph simply by re-checking them and clicking 'Redraw' again. For example, in the viewer above, it appears that recombination is occurring between the two sequences represented by the upper dark-brown and blue lines, with a recombination cross-over point around nucleotide position 5681 (on the X-axis). We would then uncheck most of the other sequences so we have only those two sequences remaining.
Once you are happy with your selection, and you decide you want to save what you've found, you may export the information in the view as a .jpg image of what you see, and/or as a text-based table of distances at each location on the X-axis. This can later be opened in Excel or similar to examine exact distances at each position.
RECOMBINATION AUTO SEARCH OPTION
The Auto Search option can be used to search through every sequence for possible recombination events. The Auto Search algorithm works on the principle that sequences contributing to recombination ('contributing sequences') will have stretches of sequence quite distant from that of the recombinant, followed or preceded by areas of relative homology to the recombinant. There are 3 parameters involved; lower threshold, upper threshold, and number of contributing sequences.
The algorithm to obtain the distance scores is the same for the single sequence viewer, but instead of only one sequence being compared to every other sequence in the alignment, every sequence is compared to every other sequence in the alignment. Using the sliding window, RAT takes the current window and the next two windows. If the genetic distance in the current window is below the lower threshold parameter and either one of the next two windows is above the upper threshold parameter, then the current sequence is flagged as a possible recombinant. For example, one may be looking at sequences that average about only 60% sequence identity, so a contributing sequence might still only have, say, 75% sequence identify with its recombinant. A search could therefore be made to look for sequences that start at less than 60% similarity to any other sequences and then rise to more than 75% similarity.
An option is provided to restrict the number of sequences allowed to contribute to recombination. In reality, it is unlikely that numerous sequences contribute to recombination in any one sequence, but as RAT searches for homology, there is a possibility that if the sequences being examined are closely related, conserved regions followed, or preceded, by areas of high variation, may be identified as areas of homology, so it is useful to restrict the number of sequences allowed to contribute to a recombination event.
The resulting output from RAT Auto Search is a JTextArea report (Fig. 4). Each sequence is named in turn, along with any evidence of recombination. If RAT finds an area of sequence that matches the input parameters, it prints out a report on the sequence, naming the contributing sequences and the region of the sequence involved in recombination. At the end of any positive sequence report, the user may view the sequences involved in the Sequence Viewer by clicking on the button provided.
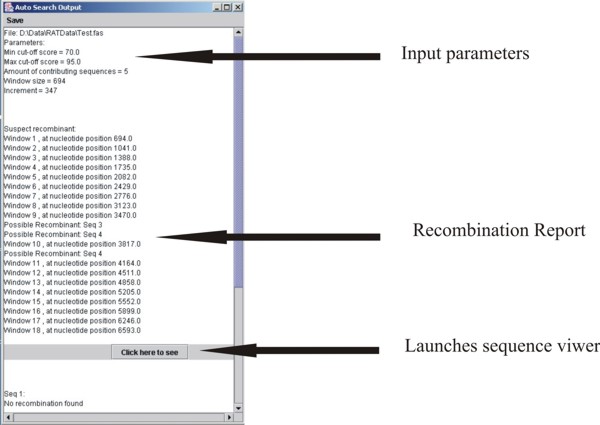 Figure 4. The Auto Search output.
Figure 4. The Auto Search output.
The view obtained is exactly that of a single sequence viewer, but with only the contributing sequences checked and displayed. It is still possible to add and remove sequences from the rest of the sequence alignment onto the graph using the check boxes in the Sequence List Pane. Using the same sequence alignment as above, it was possible to single out a recombination event and exclude all other sequences in the alignment, other than the two involved in the recombination event (below).
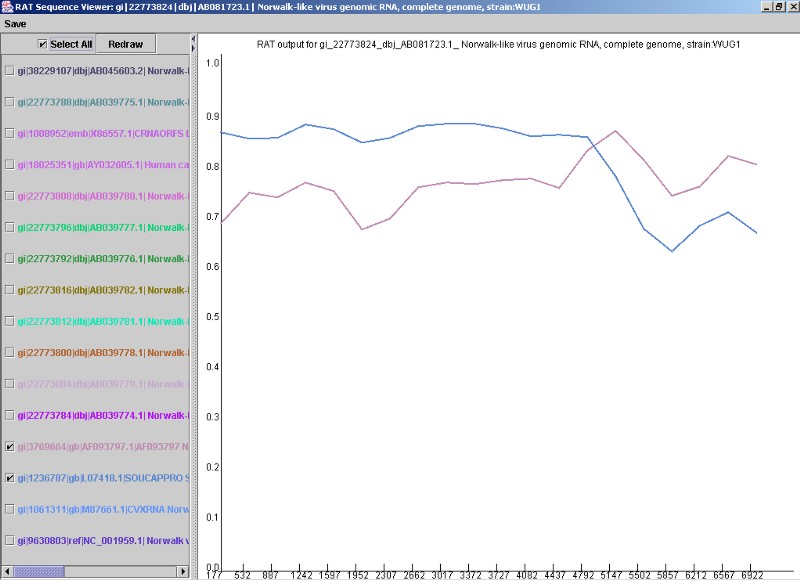 Figure 5. A recombination event found by the RAT Auto Search option.
Figure 5. A recombination event found by the RAT Auto Search option.
As the output gives the nucleotide positions of possible recombination, it is possible to narrow the search down in a further analysis by amending the 'start at position' and 'end at position' on the main GUI.
HOW 'RAT' WORKS
Single sequence viewer
The RAT algorithm is based on the distance method, using pairwise comparisons between sequences. A 'sliding window', enclosing a set number of nucleotides/amino acids, starts at the beginning of the alignment and moves along at set increments, producing a distance score for each window, repeating the distance scoring for the length of each sequence. To work out the distance score of each window, every nucleotide or amino acid of the Test Sequence in the current window is compared to each nucleotide or amino acid in the corresponding window of all other sequences in the alignment. The total number of similar sites per window are summed and then divided by the window size to give the distance score.
Auto-search
The algorithm to obtain the distance scores is the same for the single sequence viewer, but instead of only one sequence being compared to every other sequence in the alignment, every sequence is compared to every other sequence in the alignment. Using the sliding window, RAT takes the current window and the next two windows. If the current window is below the lower threshold parameter and either one of the next two windows is above the upper threshold parameter, then the current sequence is flagged as a possible recombinant. For example, one may be looking at sequences that average about only 60% sequence identity, so a contributing sequence might still only have, say, 75% sequence identify with its recombinant. A search could therefore be made to look for sequences that start at less than 60% similarity to any other sequences and then rise to more than 75% similarity.
An option is provided to restrict the number of sequences allowed to contribute to recombination. In reality, it is unlikely that numerous sequences contribute to recombination in any one sequence, but as RAT searches for similarity, there is a possibility that if the sequences being examined are closely related, conserved regions followed, or preceded, by areas of high variation, may be identified as areas of homology, so it is useful to restrict the number of sequences allowed to contribute to a recombination event.
Future versions
We would like to implement some additional features to RAT (but only as options, in order to allow users to make most use of RAT's speedy algorithm).
These include:
- Perform different types of distance analysis
- Bootscanning
- Statistical support for results