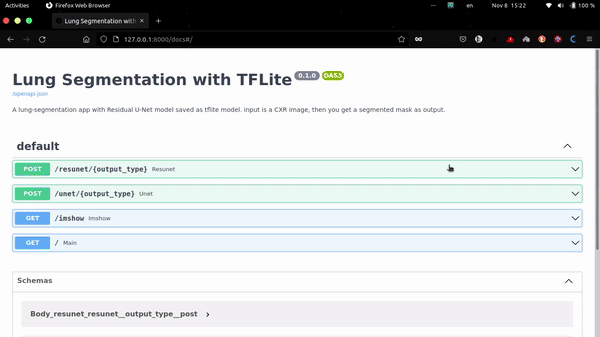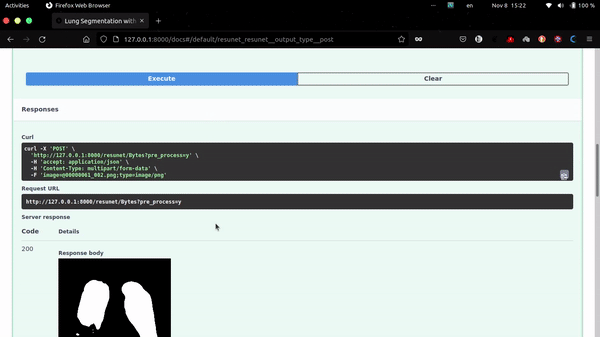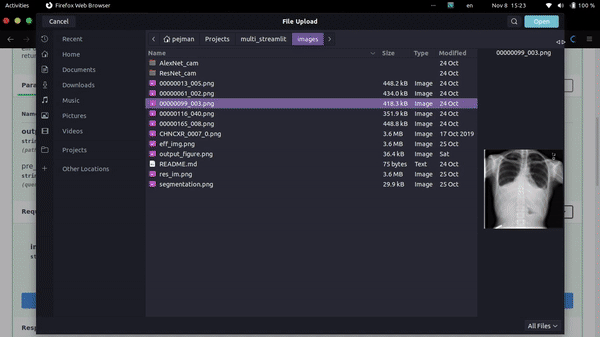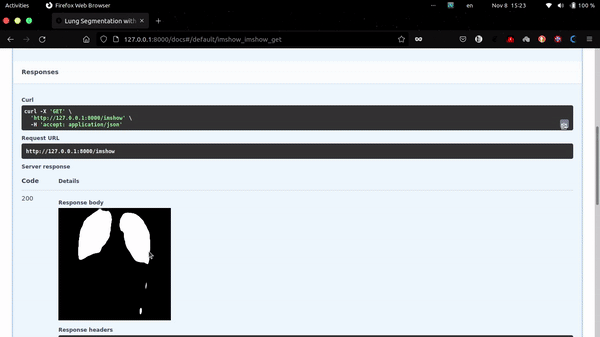
This app uses FastAPI as backend.
First install required libraries by running:
pip install -r requirements.txt
To run the application run following command in src dir:
uvicorn app:app --reload
or
chmod +x app.sh
./app.sh
http://localhost:8000/
http://localhost:8000/docs
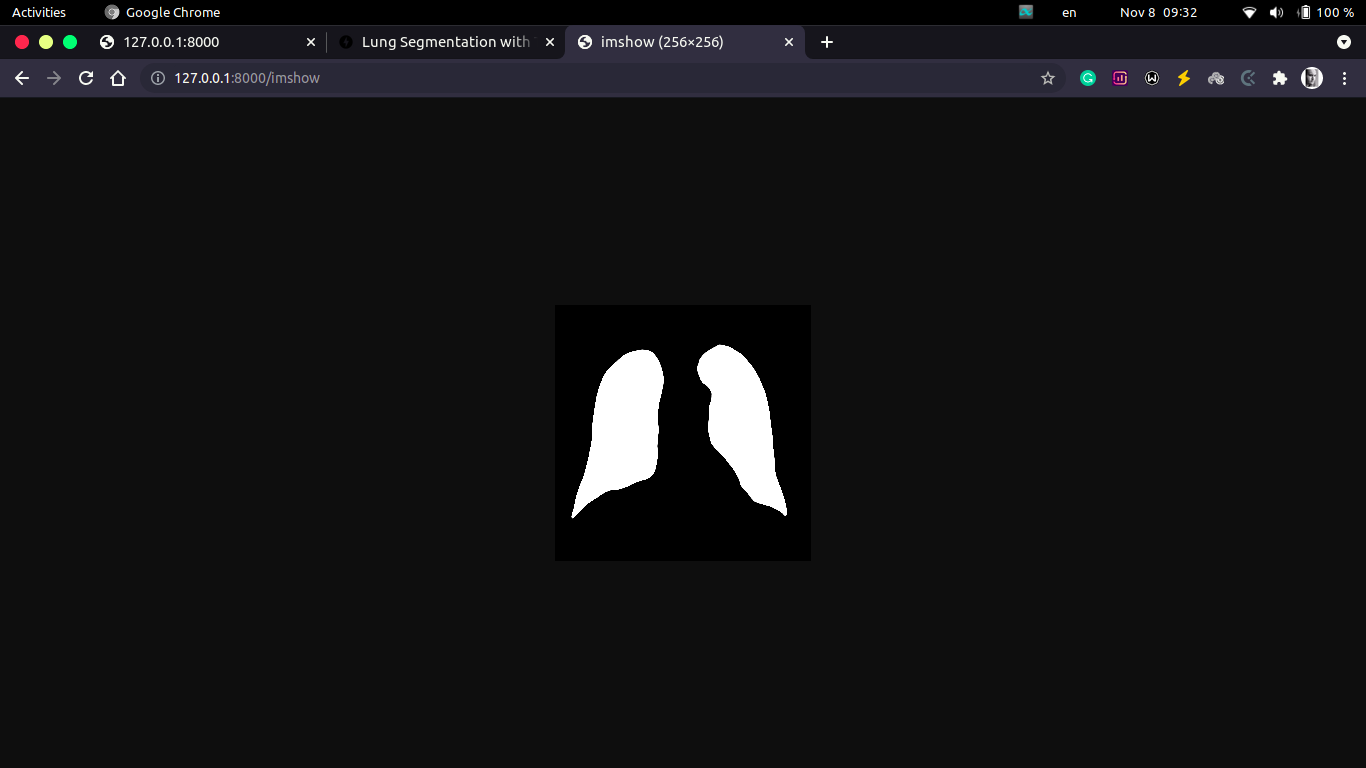
http://localhost:8000/imshow
pip install dvc dvc-gdrive
# pull weights from Google Drive
dvc pullin weights directory
weights
├── cxr_resunet.tflite
├── cxr_resunet.tflite.dvc
├── cxr_unet.tflite
└── cxr_unet.tflite.dvc
# Build image
docker build -t IMAGE_NAME:TAG_NAME .
docker run -p 8000:8000 -d IMAGE_NAME:TAG_NAMEOr
# for amd64 systems
docker run -d -p 8000:8000 pejmans21/ls-fastapi:0.1.0
#### OR
# for arm64 systems
docker run -d -p 8000:8000 pejmans21/ls-fastapi:aarch64kubectl apply -f ls-fastapi-k8s-config.yamlto see output
kubectl port-forward service/lsapi-service 8000Now check http://127.0.0.1:8000/
kubectl delete -f ls-fastapi-k8s-config.yaml