A flexible framework for rapid genome analysis and interpretation
C Chiang, R M Layer, G G Faust, M R Lindberg, D B Rose, E P Garrison, G T Marth, A R Quinlan, and I M Hall. SpeedSeq: ultra-fast personal genome analysis and interpretation. Nat Meth (2015). doi:10.1038/nmeth.3505.
http://www.nature.com/nmeth/journal/vaop/ncurrent/full/nmeth.3505.html
- Quick start
- Installation
- Reference genome and annotations
- Usage
- Example workflows
- SpeedSeq AMI (Amazon Machine Image)
- Troubleshooting
-
Install
git clone --recursive https://github.com/hall-lab/speedseq cd speedseq make -
Run the example script
cd example ./run_speedseqThis should produce the following files:
- example.bam
- example.discordants.bam
- example.splitters.bam
- example.vcf.gz
- example.sv.vcf.gz
As a template for installation on other systems, we have provided the exact commands for a full installation of SpeedSeq and GEMINI on a blank [Amazon Linux](Amazon Linux AMI 2014.09.2 (HVM)) box. These commands encompass all of the installation steps outlined below.
- g++ and the standard C and C++ development libraries (https://gcc.gnu.org/)
- CMake (http://www.cmake.org/)
- GNU awk and core utils
- Python 2.7 (https://www.python.org/)
- numpy
- pysam 0.8.0+
- scipy
- ROOT (https://root.cern.ch/) (required if running CNVnator)
- Variant Effect Predictor (http://www.ensembl.org/info/docs/tools/vep/index.html) (required if annotating VCF files)
System paths to SpeedSeq's component software are specified in the speedseq.config file, which should reside in the same directory as the SpeedSeq executable (for alternate locations use the -K flag). Upon installation, SpeedSeq attempts to automatically generate this file, but manual editing may be necessary.
The core components enable standard functionality outlined in Quick start.
- BWA (http://bio-bwa.sourceforge.net/)
- FreeBayes (https://github.com/ekg/freebayes)
- LUMPY (https://github.com/arq5x/lumpy-sv)
- Sambamba (http://lomereiter.github.io/sambamba/)
- SAMBLASTER (https://github.com/GregoryFaust/samblaster)
- Vawk (https://github.com/cc2qe/vawk)
- GNU Parallel (http://www.gnu.org/software/parallel/)
- mbuffer (http://www.maier-komor.de/mbuffer.html)
Compilation requires g++ and the standard C and C++ development libraries. Additionally, cmake is required for building the BamTools API within FreeBayes and LUMPY.
git clone --recursive https://github.com/hall-lab/speedseq
cd speedseq
make
Essential SpeedSeq components can be installed with make, which produces a log file (install.log) that details the compilation status.
The installation is modular, and its units can be built separately with make align, make var, make somatic, make sv, and make realign. This allows installation of only the desired components, eliminating extraneous dependencies. It further allows rebuilding of previously failed components.
If any components already exist on the system or fail to install, their paths can be manually specified by editing speedseq.config.
Optional components enable advanced features such as variant annotation and read-depth analysis.
- Ensembl Variant Effect Predictor (VEP) (http://www.ensembl.org/info/docs/tools/vep/index.html)
- CNVnator (http://sv.gersteinlab.org/)
curl -OL https://github.com/Ensembl/ensembl-tools/archive/release/76.zip
unzip 76.zip
perl ensembl-tools-release-76/scripts/variant_effect_predictor/INSTALL.pl \
-c $SPEEDSEQ_DIR/annotations/vep_cache \
-a ac -s homo_sapiens -y GRCh37
cp ensembl-tools-release-76/scripts/variant_effect_predictor/variant_effect_predictor.pl $SPEEDSEQ_DIR/bin
cp -r Bio $SPEEDSEQ_DIR/bin
# Update the VEP and VEP_CACHE_DIR variables in speedseq.config to point to
# $SPEEDSEQ_DIR/bin/variant_effect_predictor.pl and $SPEEDSEQ_DIR/annotations/vep_cache
CNVnator requires the ROOT package as a prerequiste (https://root.cern.ch/drupal/)
-
Install the ROOT package
curl -OL ftp://root.cern.ch/root/root_v5.34.20.source.tar.gz tar -zxvf root_v5.34.20.source.tar.gz cd root ./configure --prefix=$PWD make -
Source thisroot.sh
source /pathto/root/bin/thisroot.sh -
Compile CNVnator from the SpeedSeq directory
cd $SPEEDSEQ_DIR make cnvnator -
Before running SpeedSeq, you'll need to add the following line to speedseq.config or your .bashrc file. (Substitute the actual path to thisroot.sh on your system)
source /pathto/root/bin/thisroot.sh
Please refer to the CNVnator repository for details on installing CNVnator.
We recommend using the GRCh37 human genome for SpeedSeq, available here:
ftp://ftp-trace.ncbi.nih.gov/1000genomes/ftp/technical/reference/human_g1k_v37.fasta.gz
ftp://ftp-trace.ncbi.nih.gov/1000genomes/ftp/technical/reference/human_g1k_v37.fasta.fai
The genome FASTA file should be unzipped and indexed with BWA before running SpeedSeq.
For human genome alignment using the GRCh37 build, we recommend using the annotations/ceph18.b37.include.2014-01-15.bed windows to parallelize variant calling (speedseq var and speedseq somatic). This BED file excludes 15.6 Mb of the non-gapped genome where the coverage in the CEPH1463 pedigree was greater than twice the mode coverage plus 3 standard deviations. We believe these extremely high depth regions that we excluded are areas of misassembly in the GRCh37 human reference genome in which variant calling is time-consuming and error-prone.
Additionally, the regions in annotations/ceph18.b37.include.2014-01-15.bed are variable-width windows which each contain approximately the same coverage depth in the CEPH1463 pedigree, and sorted from highest to lowest depth. This ensures that the parallelization of Freebayes uses approximately the same amount of time per region.
The regions in annotations/ceph18.b37.exclude.2014-01-15.bed represent the complement of the regions in annotations/ceph18.b37.include.2014-01-15.bed.
In the speedseq sv module, we recommend excluding the genomic regions in the annotations/ceph18.b37.lumpy.exclude.2014-01-15.bed BED file. These regions represent the complement of those in annotations/ceph18.b37.include.2014-01-15.bed as well as the mitochondrial chromosome.
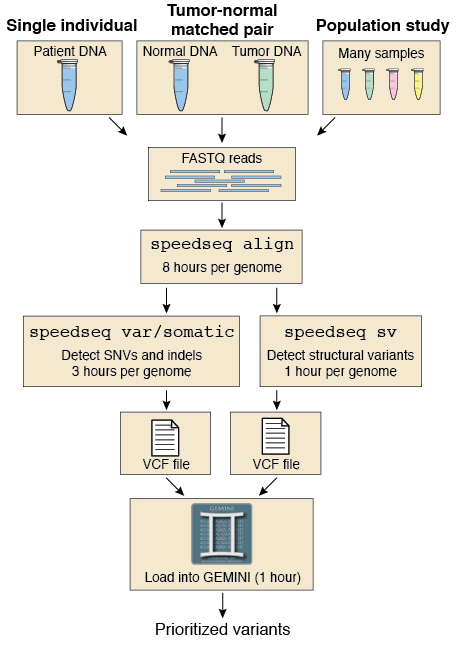
SpeedSeq is a modular framework with four components:
- speedseq align - Process paired-end FASTQ sequences to produce a duplicate-marked, sorted, indexed BAM file that can be processed with other SpeedSeq modules.
- speedseq var - Run FreeBayes one or more BAM files
- speedseq somatic - Run FreeBayes on a tumor/normal pair of BAM files
- speedseq sv - Run LUMPY on one or more BAM files, with optional breakend genotyping and read-depth calculation.
- speedseq realign - Realign from a BAM file.
These modules operate independently of each other and produce universal output formats that are compatible with external tools. SpeedSeq modules can also run on BAM alignments that were produced outside of the SpeedSeq framework. . However, structural variant detection on BAM files generated outside of SpeedSeq will be slower due to two unique features of speedseq align. First, our alignment uses SAMBLASTER to automatically extract split and discordant reads for SV detection. While the speedseq sv module will internally extract split and discordant reads from regular BAM files, it takes much longer due to obligate name-sorting of the BAM file. Secondly, structural variant genotyping is much faster on BAM files processed by SAMBLASTER due to the addition of mate CIGAR and mate mapping quality tags. In the absence of these tags, SVTyper must jump to each read’s mate position in the BAM file, which greatly increases run time.
speedseq align converts paired-end FASTQ sequences to a duplicate-marked, sorted, indexed BAM file that can be processed with other SpeedSeq modules.
Internally, speedseq align runs the following steps to produce three output BAM files:
- Alignment with BWA-MEM
- Duplicate marking with SAMBLASTER
- Discordant-read and split-read extraction with SAMBLASTER
- Position sorting with Sambamba
- BAM indexing with Sambamba
usage: speedseq align [options] <reference.fa> <in1.fq> [in2.fq]
reference.fa genome reference fasta file (required)
in1.fq paired-end fastq file. if -p flag is used then expected to be
an interleaved paired-end fastq file, and in2.fq may be omitted.
(may be gzipped) (required)
in2.fq paired-end fastq file. (may be gzipped) (required)
-o STR output prefix [default: in1.fq]
-R read group header line such as "@RG\tID:id\tSM:samplename\tLB:lib" (required)
-p first fastq file consists of interleaved paired-end sequences
-t INT number of threads to use [default: 1]
-T DIR temp directory [./outprefix.XXXXXXXXXXXX]
-I FLOAT[,FLOAT[,INT[,INT]]]
specify the mean, standard deviation (10% of the mean if absent), max
(4 sigma from the mean if absent) and min of the insert size distribution.
FR orientation only. [inferred]
-i include duplicates in splitters and discordants
(default: exclude duplicates)
-c INT maximum number of split alignments for a read to be
included in splitter file [default: 2]
-m INT minimum non-overlapping base pairs between two alignments
for a read to be included in splitter file [default: 20]
-M amount of memory in GB to be used for sorting [default: 20]
-K FILE path to speedseq.config file (default: same directory as speedseq)
-v verbose
-h show help message
speedseq align produces three sorted, indexed BAM files (plus their corresponding .bai index files):
outprefix.bam- The full, duplicate-marked, sorted BAM file for the library. This file may serve as input for
speedseq var,speedseq somatic, andspeedseq sv.
- The full, duplicate-marked, sorted BAM file for the library. This file may serve as input for
outprefix.splitters.bam- This BAM file contains split reads called by the BWA-MEM alignment of the library. It may be used as the
-Sflag input tospeedseq sv. This file excludes duplicate reads by default, but they will be included if the-iflag is specified as aspeedseq aligncommand line parameter.
- This BAM file contains split reads called by the BWA-MEM alignment of the library. It may be used as the
outprefix.discordants.bam- This BAM file contains discordant read-pairs called by the BWA-MEM alignment of the library. These reads may be discordant by strand orientation, intrachromosomal distance, or interchromosomal mapping. This BAM file may be used as the
-Dflag input tospeedseq sv. This file excludes duplicate reads by default, but they will be included if the-iflag is specified as aspeedseq aligncommand line parameter.
- This BAM file contains discordant read-pairs called by the BWA-MEM alignment of the library. These reads may be discordant by strand orientation, intrachromosomal distance, or interchromosomal mapping. This BAM file may be used as the
speedseq var runs FreeBayes on one or more BAM files.
usage: speedseq var [options] <reference.fa> <input1.bam> [input2.bam [...]]
reference.fa genome reference fasta file
input.bam BAM file(s) to call variants on. Must have readgroup information,
and the SM readgroup tags will be the VCF column headers
-o STR output prefix [default: input1.bam]
-w FILE BED file of windowed genomic intervals. For human genomes,
we recommend using the annotations/ceph18.b37.include.2014-01-15.bed
(see Annotations)
-q FLOAT minimum variant QUAL score to output [1]
-t INT number of threads to use [default: 1]
-T DIR temp directory [./outprefix.XXXXXXXXXXXX]
-A annotate the vcf with VEP
-K FILE path to speedseq.config file [default: same directory as speedseq]
-v verbose
-h show help message
speedseq var produces a single indexed VCF file that is optionally annotated with VEP.
outprefix.vcf.gz
speedseq somatic runs FreeBayes on a tumor/normal pair of BAM files
usage: speedseq somatic [options] <reference.fa> <normal.bam> <tumor.bam>
reference.fa genome reference fasta file
normal.bam germline BAM file(s) (comma separated BAMs from multiple libraries).
Must have readgroup information, and the SM readgroup tag will
be the VCF column header
tumor.bam tumor BAM file(s) (comma separated BAMs for multiple libraries).
Must have readgroup information, and the SM readgroup tag will
be the VCF column header
-o STR output prefix [default: tumor.bam]
-w FILE BED file of windowed genomic intervals. For human genomes,
we recommend using the annotations/ceph18.b37.include.2014-01-15.bed
(see Annotations)
-t INT number of threads to use [default: 1]
-F FLOAT require at least this fraction of observations supporting
an alternate allele within a single individual in order
to evaluate the position [0.05]
-C INT require at least this count of observations supporting
an alternate allele within a single individual in order
to evaluate the position [2]
-S FLOAT minimum somatic score (SSC) for PASS [18]
-q FLOAT minimum QUAL score to output non-passing somatic variants [1e-5]
-T DIR temp directory [./outprefix.XXXXXXXXXXXX]
-A annotate the vcf with VEP
-K FILE path to speedseq.config file (default: same directory as speedseq)
-v verbose
-h show help message
speedseq somatic produces a single indexed VCF file that is optionally annotated with VEP.
outprefix.vcf.gz
speedseq sv runs LUMPY on one or more BAM files, with optional breakend genotyping by SVTyper, and optional read-depth analysis by CNVnator.
-B FILE full BAM file(s) (comma separated) (required)
example: -B in1.bam,in2.bam,in3.bam
-S FILE split reads BAM file(s) (comma separated, order same as in -B)
(auto-generated if absent)
example: -S in1.splitters.bam,in2.splitters.bam,in3.splitters.bam
-D FILE discordant reads BAM file(s) (comma separated, order same as in -B)
(auto-generated if absent)
example: -D in1.discordants.bam,in2.discordants.bam,in3.discordants.bam
-R FILE indexed reference genome fasta file (required)
-o STR output prefix [in1.bam]
-t INT threads [1]
-x FILE BED file to exclude
-g genotype SV breakends with svtyper
-d calculate read-depth with CNVnator
-A annotate the vcf with VEP
-P output LUMPY probability curves in VCF
-m INT minimum sample weight for a call [default: 4]
-r FLOAT trim threshold [0]
-T DIR temp directory [./outprefix.XXXXXXXXXXXX]
-k keep temporary files
-K FILE path to speedseq.config file (default: same directory as speedseq)
-v verbose
-h show help message
speedseq sv produces a bgzipped, indexed VCF file.
outprefix.sv.vcf.gz
speedseq realign allows alignment from one or more BAM files, rather than FASTQ inputs. It automatically parses read group information from the BAM header to mark duplicates by library.
usage: speedseq realign [options] <reference.fa> <in1.bam> [in2.bam [...]]
reference.fa genome reference fasta file (indexed with bwa)
in.bam BAM file(s) (must contain read group tags)
-o STR output prefix [in.realign]
-I FLOAT[,FLOAT[,INT[,INT]]]
specify the mean, standard deviation (10% of the mean if absent), max
(4 sigma from the mean if absent) and min of the insert size distribution.
FR orientation only. [inferred]
-n rename reads for smaller file size
-t INT threads [1]
-T DIR temp directory [./output_prefix.XXXXXXXXXXXX]
-i include duplicates in splitters and discordants
(default: exclude duplicates)
-c INT maximum number of split alignments for a read to be
included in splitter file [default: 2]
-m INT minimum non-overlapping base pairs between two alignments
for a read to be included in splitter file [default: 20]
-M amount of memory in GB to be used for sorting [default: 20]
-K FILE path to speedseq.config file (default: same directory as speedseq)
-v verbose
-h show help message
speedseq realign output is identical to that produced by speedseq align.
-
Use
speedseq alignto produce a sorted, duplicate-marked, BAM alignment from paired-end fastq data.speedseq align \ -o NA12878 \ -R "@RG\tID:NA12878.S1\tSM:NA12878\tLB:lib1" \ human_g1k_v37.fasta \ NA12878.1.fq.gz \ NA12878.2.fq.gzNote: if using an interleaved paired-end fastq file, use the
-pflagspeedseq align \ -o NA12878 \ -p \ -R "@RG\tID:NA12878.S1\tSM:NA12878\tLB:lib1" \ human_g1k_v37.fasta \ NA12878.interleaved.fq.gz -
Use
speedseq varto call SNVs and indels on a single sample.speedseq var \ -o NA12878 \ -w annotations/ceph18.b37.include.2014-01-15.bed \ human_g1k_v37.fasta \ NA12878.bam -
Use
speedseq svto call structural variants. The optional-gand-dflags perform breakend genotyping and read-depth calculation respectivelyspeedseq sv \ -o NA12878 \ -x annotations/ceph18.b37.lumpy.exclude.2014-01-15.bed \ -g \ -d \ -B NA12878.bam \ -D NA12878.discordants.bam \ -S NA12878.splitters.bam
-
Use
speedseq alignto produce a sorted, duplicate-marked, BAM alignment of each library.speedseq align -o NA12878_S1 -R "@RG\tID:NA12878.S1\tSM:NA12878\tLB:lib1" \ human_g1k_v37.fasta \ NA12878.S1.1.fq.gz \ NA12878.S1.2.fq.gz speedseq align -o NA12878_S2 -R "@RG\tID:NA12878.S2\tSM:NA12878\tLB:lib2" \ human_g1k_v37.fasta \ NA12878.S2.1.fq.gz \ NA12878.S2.2.fq.gz speedseq align -o NA12878_S3 -R "@RG\tID:NA12878.S3\tSM:NA12878\tLB:lib3" \ human_g1k_v37.fasta \ NA12878.S3.1.fq.gz \ NA12878.S3.2.fq.gz -
Merge the samples
sambamba merge NA12878_merged.bam NA12878_S1.bam NA12878_S2.bam NA12878_S3.bam sambamba index NA12878_merged.bam -
Use
speedseq varto call SNVs and indels.speedseq var \ -o NA12878 \ -w annotations/ceph18.b37.include.2014-01-15.bed \ human_g1k_v37.fasta \ NA12878_merged.bam -
Use
speedseq svto call structural variants.speedseq -sv \ -o NA12878 \ -x annotations/ceph18.b37.lumpy.exclude.2014-01-15.bed \ -B NA12878_merged.bam \ -S NA12878_merged.splitters.bam \ -D NA12878_merged.discordants.bam \ -R human_g1k_v37.fasta
-
Use
speedseq alignto produce sorted, duplicate-marked, BAM alignments for each sample.speedseq align \ -o NA12877 \ -R "@RG\tID:NA12877.S1\tSM:NA12877\tLB:lib1" \ human_g1k_v37.fasta \ NA12877.1.fq.gz \ NA12877.2.fq.gz speedseq align \ -o NA12878 \ -R "@RG\tID:NA12878.S1\tSM:NA12878\tLB:lib2" \ human_g1k_v37.fasta \ NA12878.1.fq.gz \ NA12878.2.fq.gz speedseq align \ -o NA12879 \ -R "@RG\tID:NA12879.S1\tSM:NA12879\tLB:lib3" \ human_g1k_v37.fasta \ NA12879.1.fq.gz \ NA12879.2.fq.gz -
Use
speedseq varto call SNVs and indels on multiple samples.speedseq var \ -o cephtrio \ -w annotations/ceph18.b37.include.2014-01-15.bed \ human_g1k_v37.fasta \ NA12877.bam \ NA12878.bam \ NA12879.bam -
Use
speedseq svto call structural variants on multiple samples.speedseq sv \ -o cephtrio \ -x annotations/ceph18.b37.lumpy.exclude.2014-01-15.bed \ -B NA12877.bam,NA12878.bam,NA12879.bam \ -D NA12877.discordants.bam,NA12878.discordants.bam,NA12879.discordants.bam \ -S NA12877.splitters.bam,NA12878.splitters.bam,NA12879.splitters.bam
-
Use
speedseq alignto produce sorted, duplicate-marked, BAM alignments for the tumor/normal pairspeedseq align \ -o TCGA-B6-A0I6.normal \ -p \ -R "@RG\tID:TCGA-B6-A0I6-10A-01D-A128-09\tSM:TCGA-B6-A0I6-10A-01D-A128-09\tLB:lib1" \ human_g1k_v37.fasta \ TCGA-B6-A0I6-10A-01D-A128-09.interleaved.fq.gz speedseq align \ -o TCGA-B6-A0I6.tumor \ -p \ -R "@RG\tID:TCGA-B6-A0I6-10A-01D-A128-09\tSM:TCGA-B6-A0I6-10A-01D-A128-09\tLB:lib1" \ human_g1k_v37.fasta \ TCGA-B6-A0I6-01A-11D-A128-09.interleaved.fq.gz -
Use
speedseq somaticto call SNVs and indels on the tumor/normal pair.speedseq somatic \ -o TCGA-B6-A0I6 \ -w annotations/ceph18.b37.include.2014-01-15.bed \ -F 0.05 \ -q 1 \ human_g1k_v37.fasta \ TCGA-B6-A0I6.normal.bam \ TCGA-B6-A0I6.tumor.bam -
Use
speedseq svto call structural variants on the tumor/normal pair.speedseq sv \ -o TCGA-B6-A0I6 \ -x annotations/ceph18.b37.lumpy.exclude.2014-01-15.bed \ -B TCGA-B6-A0I6.normal.bam,TCGA-B6-A0I6.tumor.bam \ -D TCGA-B6-A0I6.normal.discordants.bam,TCGA-B6-A0I6.tumor.discordants.bam \ -S TCGA-B6-A0I6.normal.splitters.bam,TCGA-B6-A0I6.tumor.splitters.bam \ -R human_g1k_v37.fasta
-
Use
speedseq alignto produce sorted, duplicate-marked, BAM alignments (as shown above). -
Use
speedseq varto call SNVs and indels on multiple samples with high sensitivity.speedseq var \ -o trio \ -w annotations/ceph18.b37.include.2014-01-15.bed \ -q 0.01 \ human_g1k_v37.fasta \ mother.bam \ father.bam \ child.bam -
Filter variants for de novo status using GEMINI's built-in analysis tools.
SpeedSeq is available as a public AMI (Amazon Machine Image) on the Amazon Elastic Compute Cloud (EC2).
-
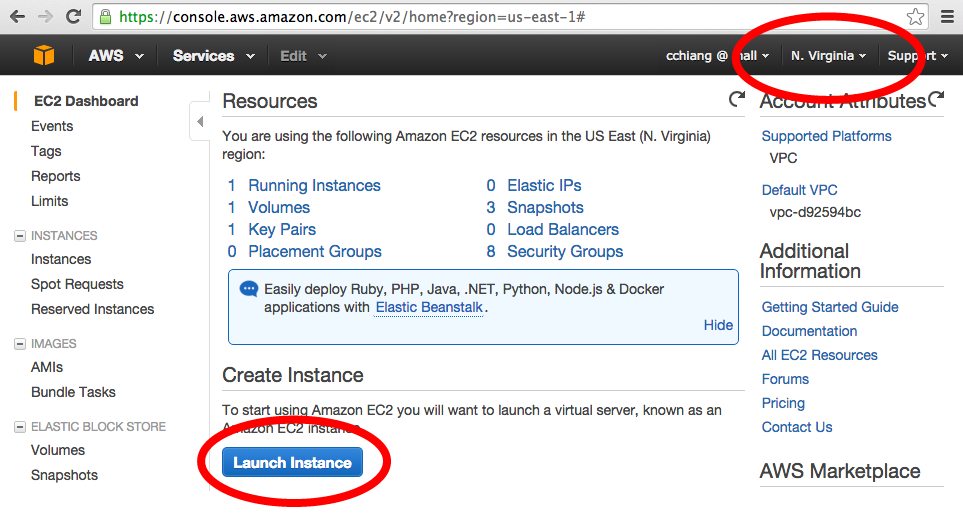
Log in to the Amazon AWS console
-
From the EC2 Dashboard, set your region to N. Virginia and click "Launch Instance"
-
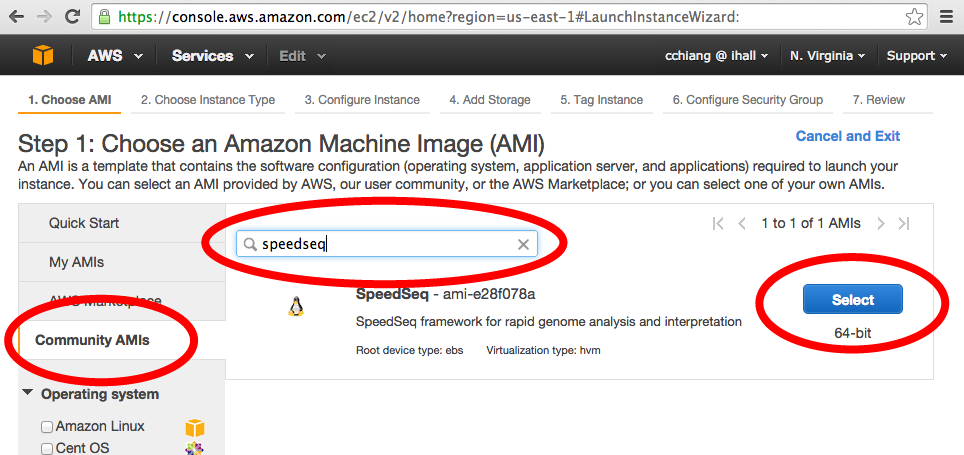
Choose, "Community AMIs" in the sidebar and search for SpeedSeq in the dialog box.
-
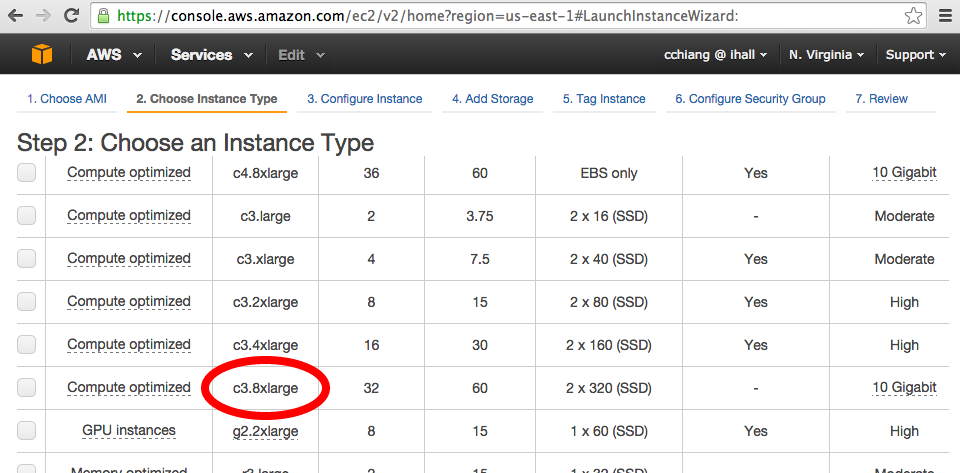
Select hardware specifications. For deep whole genomes, we recommend c3.8xlarge (32 vCPUs, 60 GB RAM). For testing purposes any machine with 16 GB RAM is sufficient.
-
Add storage for the data. Note that the SpeedSeq footprint is ~ 26 GB (including the reference genome and GEMINI).
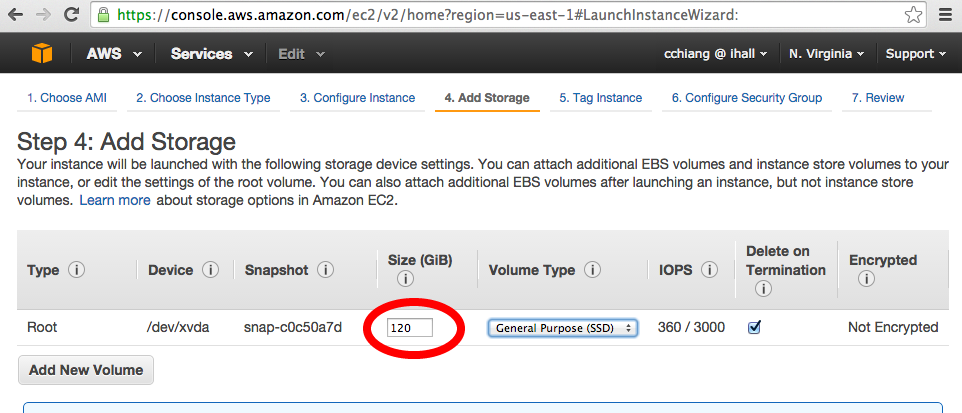
-
Launch the instance and log in.
ssh -i mykey.pem ec2-user@ec2-54-173-62-218.compute-1.amazonaws.com -
Run the SpeedSeq test script.
./run_speedseq.shThis should produce the following files:
- example.bam
- example.discordants.bam
- example.splitters.bam
- example.vcf.gz
- example.sv.vcf.gz
- Can I use SpeedSeq on exome data?
- SpeedSeq can detect SNVs and indels from exome data using
speedseq varorspeedseq somatic. However, you should not use the excluded regions, as these were designed for WGS data. SpeedSeq cannot detect SVs from exome data.
- SpeedSeq can detect SNVs and indels from exome data using
If you encounter errors or strange behavior from SpeedSeq, please report them to the issues page with the following information:
- Description of the problem
- The exact command that you ran, using the "-v" option for verbose logging information
- Any error or status information produced by SpeedSeq
- Anything you've tried to resolve the issue
- Installation failure with error: "No targets specified and no makefile found."
Ensure that SpeedSeq was cloned with the
--recursiveflag
- Installation failure while compiling FreeBayes or LUMPY (in the var and sv modules respectively)
These two components use BamTools, which requires CMake for compilation. Ensure that CMake is installed on your system
- Installation reports, "WARNING: CNVnator not compiled because the ROOT package is not installed. Please see the README for instructions on manually installing ROOT."
This indicates that the ROOT package has not been installed, or the $ROOTSYS variable has not been set. See the CNVnator repository for details.
- Runtime error: "ImportError: No module named argparse"
Ensure you are running Python 2.7 or later.
- CNVnator fails to run during
speedseq sv
This may be due to a problem with the ROOT installation. Try configuring the ROOT package without the --prefix flag. Then run
./configure make
Add /pathto/root/bin/thisroot.sh to the speedseq.config file
- Python errors
Python errors commonly result from incompatibilities with older versions of Pysam. SpeedSeq runs on Pysam versions 0.8.0 and newer.