Installing is as always
pip install benchmcmc
It depends on pymc3 and
matplotlib, so these are also
installed.
Getting started real quick:
benchmcmc --generate 69 11 10 131 10 9 --beta > bench.txt
benchmcmc bench.txt
The first command generates 200 synthetic data points and put them into
bench.txt, and the second command runs benchmcmc on the data,
displaying a trace plot of the MCMC.
This package lets you take a series of benchmark data analyses whether there at one point was a change in performance.
Suppose that you run your benchmark tests on every commit that you have
(e.g. looping over git-rev-list),
and you see that your performance data
is (e.g. in requests per second or in seconds, or other measures)
as follows:
13.64
12.82
11.69
15.12
12.30
18.46
13.51
14.33
13.84
12.77
... (180 rows omitted)
10.93
11.02
12.45
11.78
12.12
13.51
10.66
10.18
10.81
12.19
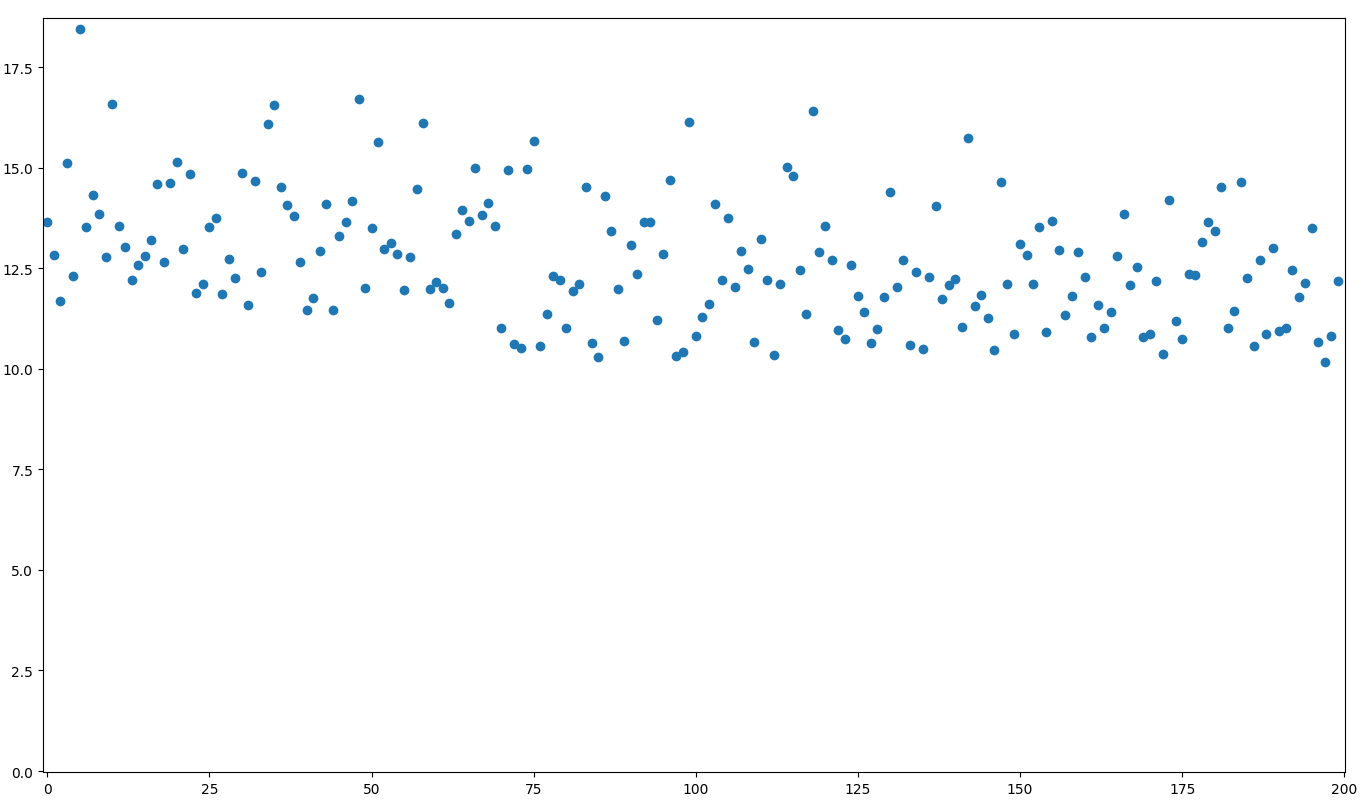
In this data, it seems to be centered around 13+ε in the beginning, and it ends being centered around 12+ε, or visualized:
There seems to be a slight drop in values before the 100th point, but it's not easy to determine exactly where the change occurred.
Suppose that you wonder whether or not the performance at the start and at the end are likely to be from two different distributions, and if so, where the switchpoint was.
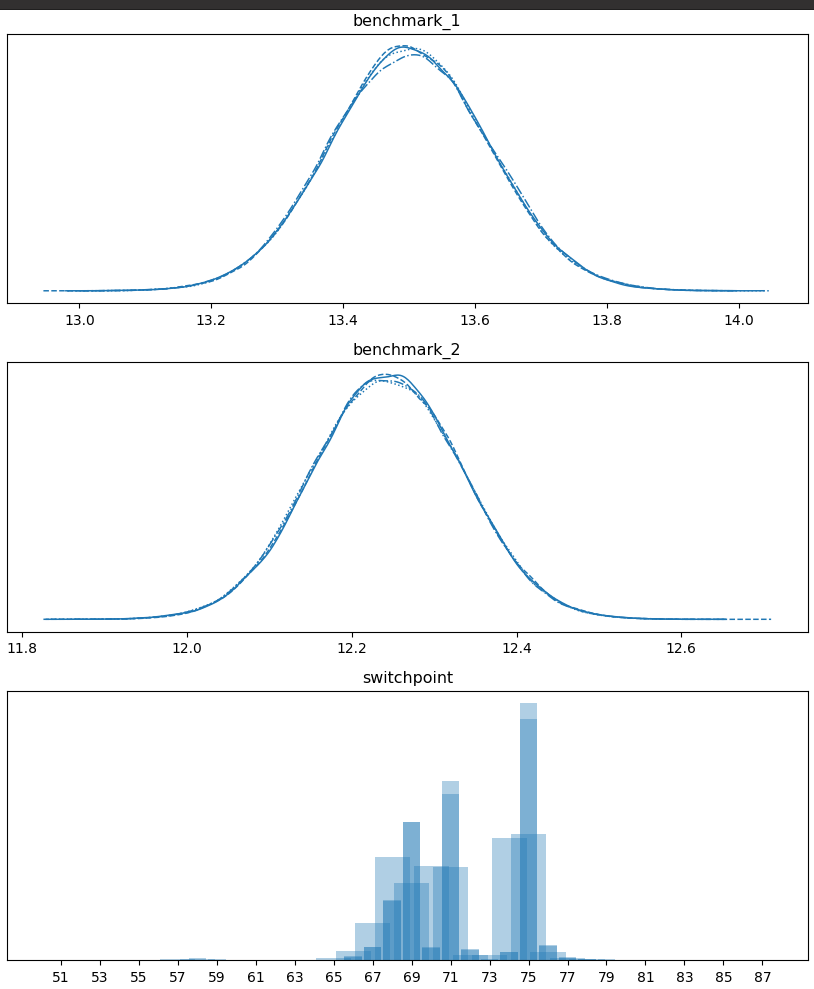
Running benchmcmc on the data gives the above plot which shows that it
is likely that the performance went from ~13.5 to ~12.25 at or around the
69th or 75th datapoints.
This helps you pin down when a performance change might have occurred.
$ benchmcmc bench.txt --draws 1000 --tune 1000 --cores 4 --target-accept 0.8
The options available are those illustrated in the above examples and
all taken from and fed into
pymc3.sampling.sample.
draws— The number of samples to draw. Defaults to 1000. The number of tuned samples are discarded by default.tune— Number of iterations to tune, defaults to 1000. Samplers adjust the step sizes, scalings or similar during tuning. Tuning samples will be drawn in addition to the number specified in the draws argument, and will be discarded.cores— The number of chains to run in parallel. IfNone, set to the number of CPUs in the system, but at most 4.target_accept: float in [0, 1] — The step size is tuned such that we approximate this acceptance rate. Higher values like 0.9 or 0.95 often work better for problematic posteriors.
You can run benchmcmc --generate for generating synthetic benchmark
data.
$ benchmcmc --generate 100 15 3 100 14 3 [--beta] > benchmarkfile.txtThis generates 200 samples, 100 from N(mu=15, sigma=3) followed by 100
from N(mu=14, sigma=3).
If you use --beta, you get a bit more realistic performance with a
lower bound of mu, especially for lower values of mu.
Suppose that you want to run python script.py on a script that is in
your Git tree.
LOGFILE=/tmp/timescript
echo "" > $LOGFILE
for commit in $(git rev-list master)
do
git checkout $commit
printf "%s," "`(git rev-parse --short HEAD)`" >> $LOGFILE
/usr/bin/time -a -o $LOGFILE --format=%e python script.py
done
tac $LOGFILEWhen run in a repository, it will output time data in the format
commit,time
Here is an example of the output:
484fde8,0.04
58a1cdb,0.04
d26b797,0.04
81f4b9a,0.04
3ae1e11,0.04
7689ca2,0.04
8c76b29,0.04
43db50c,0.04
b34b146,0.04
4c56a54,0.04
9c08050,0.07
b22278d,0.07
7a9c111,0.07
065b6a5,0.07
6cc7cdd,0.07
ec7f042,0.07
b3ba887,0.08
a32ce81,0.07
9136914,0.07
b456714,0.07
504cf73,0.07
8002774,0.07
e1f5f9f,0.09
For example, you can create a script gitrun that looks as follows:
#!/usr/bin/bash
GITRUN_EXEC=$1
shift
GITRUN_ARGS=$@
for commit in $(git rev-list master)
do
git checkout --quiet $commit
printf "%s," "`(git rev-parse --short HEAD)`"
sh -c "$GITRUN_EXEC $GITRUN_ARGS"
done