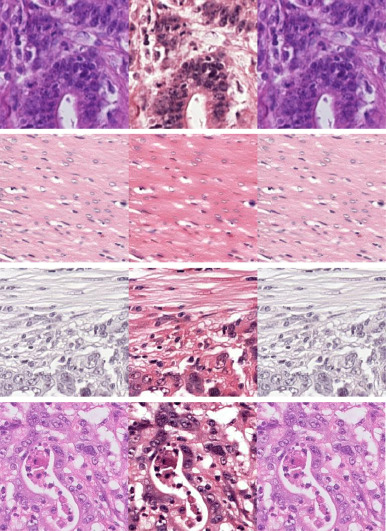
This repository implements the model described in 'Residual cyclegan for robust domain transformation of histopathological tissue slides'. The paper builds further on the original CycleGAN approach.
The original repository does not provide all necessary internal functions to train a residual cycle GAN. In this repository, I am adding these functions to enable model training and prediction.
The purpose of this implementation is two-fold:
- To transform low-resolution images of IHC WSIs to H&E in order to apply an H&E tissue detection algorithm. This approach is inspired by de Vulpian et al. (https://2022.midl.io/papers/d_s_14).
- To transform tiles between the IHC and H&E domain at full resolution.
The tissue detection pipeline will be fully developed using training data from the ACROBAT challenge (https://acrobat.grand-challenge.org/).