Transcriptome-wide prediction of prostate cancer gene expression from histopathology images using co-expression based convolutional neural networks
This repository showcases how to efficiently perform a transcriptome-wide analysis of gene expression prediction with convolutional neural networks by exploiting the co-expression of transcripts. This example relies entirely on publicly available data from the GDC data portal of the TCGA research consortium.
Molecular phenotyping by gene expression profiling is central in contemporary cancer research and in molecular diagnostics but remains resource intense to implement. Changes in gene expression occurring in tumours cause morphological changes in tissue, which can be observed on the microscopic level. The relationship between morphological patterns and some of the molecular phenotypes can be exploited to predict molecular phenotypes from routine haematoxylin and eosin (H&E) stained whole slide images (WSIs) using convolutional neural networks (CNNs). In this study, we propose a new, computationally efficient approach to model relationships between morphology and gene expression. We conducted the first transcriptome-wide analysis in prostate cancer, using CNNs to predict bulk RNA-sequencing estimates from WSIs for 370 patients from the TCGA PRAD study. Out of 15586 protein coding transcripts, 6618 had predicted expression significantly associated with RNA-seq estimates (FDR-adjusted p-value < 1*10-4) in a cross-validation. 5419 (81.9%) of these associations were subsequently validated in a held-out test set. We furthermore predicted the prognostic cell cycle progression score directly from WSIs. These findings suggest that contemporary computer vision models offer an inexpensive and scalable solution for prediction of gene expression phenotypes directly from WSIs, providing opportunity for cost-effective large-scale research studies and molecular diagnostics.
A draft of our manuscript is available from arxiv.org: https://arxiv.org/abs/2104.09310
This example uses the same patient selection and data split as the publication:
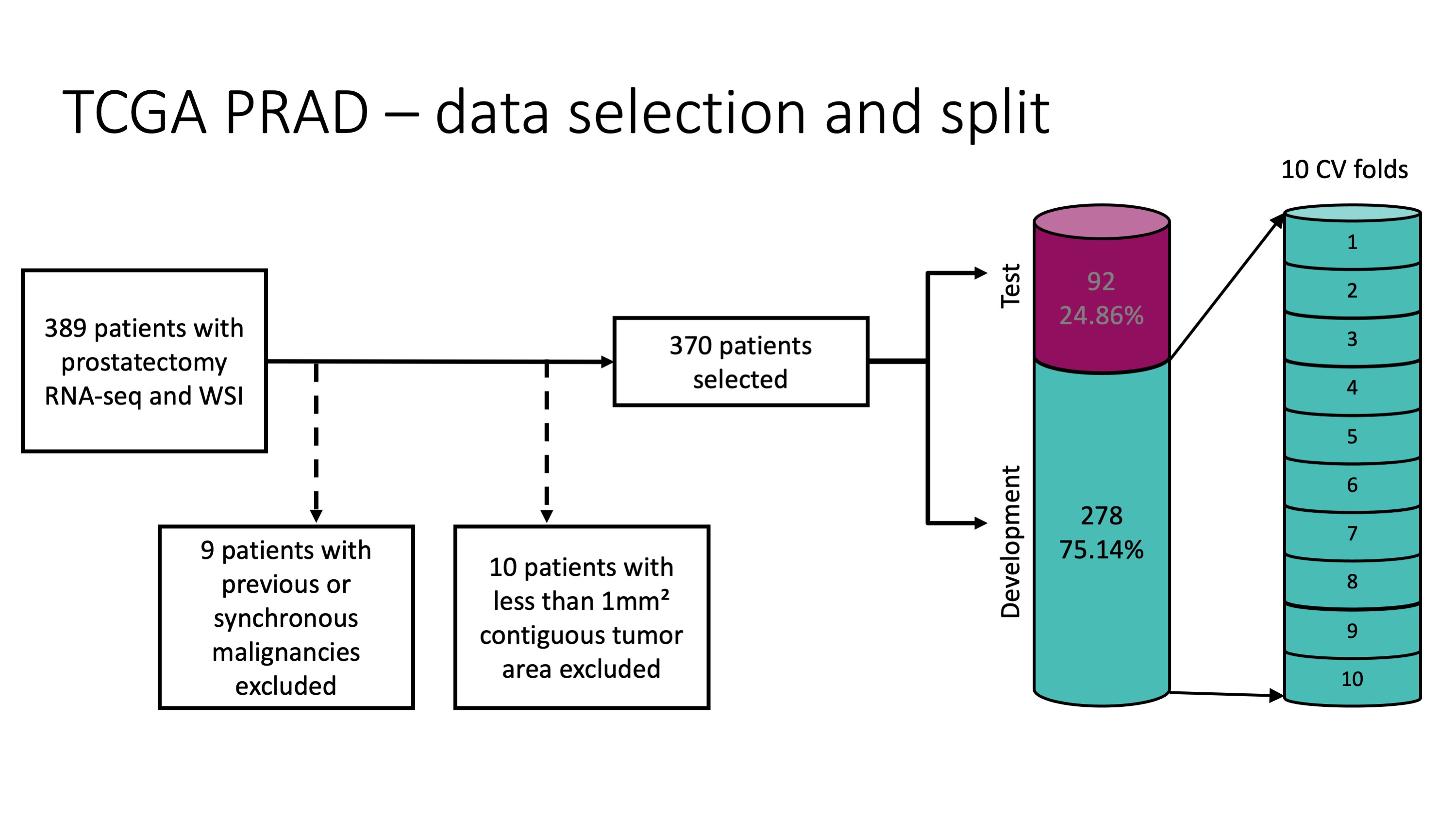
Genes are selected and predicted as depicted in this schematic:
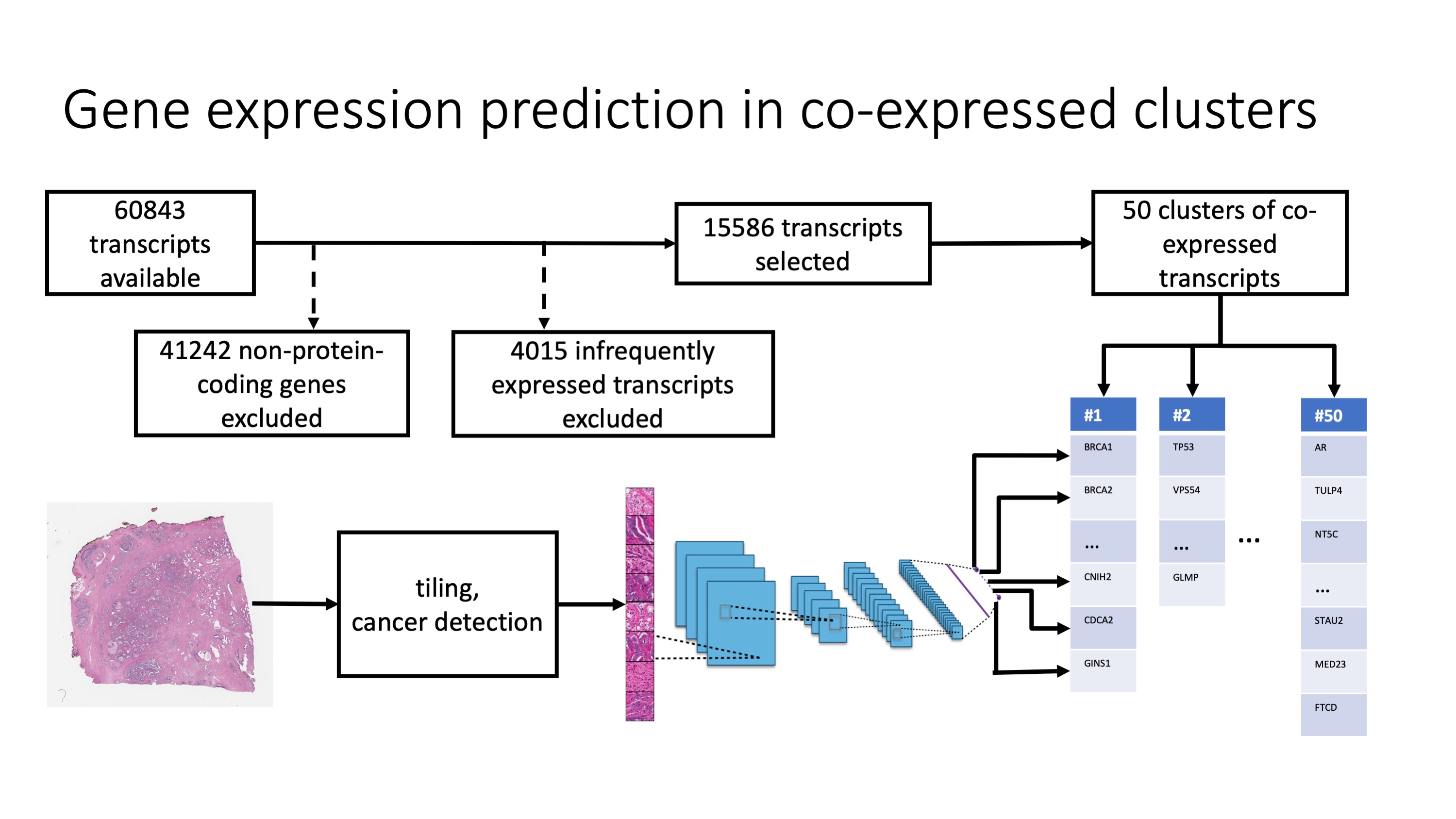
-
Download the GDC Data Transfer Tool that is appropriate for your OS. Now unzip the gdc client and grant permissions to it by typing
chmod u+x gdc-client. -
Create a directory to which the slides will be downloaded. This will require approximately 300GB of free disk space. Copy the gdc client to this directory, along with the file
manifest_wsi.txt, to be found underdatain this repository. Now run the command./gdc-client download -m manifest_wsi.txtin a terminal. This parent directory that contains the folders with the.svsfiles of the slides will be referred to aspath/to/slidesfrom here on. -
Create a second directory to whith the RNA-seq data will be downloaded. Move the gdc-client to this directory and copy the
manifest_rna_seq_fpkm_uq.txtto this directory. Download the RNA-seq data by executing./gdc-client download -m manifest_rna_seq_fpkm_uq.txt. The folder cotaining the RNA-seq data will be referred to aspath/to/rna-seq.
Once the RNA-seq data is downloaded, create a pandas data frame with the RNA labels for model training by navigating to the folder 1_data_preparation and running
python get_df_rna.py --indir path/to/rna-seq
This will save a data frame with RNA-seq labels for model fitting under data.
WSIs are too large to fit into GPU memory. One way to deal with this is to divide WSIs into smaller image patches and assign the slide-level label to each image patch as a weak label. In order to tile the downloaded wsis, navigate to the folder 1_data_preparation and run
python get_df_rna.py --indir path/to/slides --outdir path/to/tiles --n_jobs 1
This will use the coordinates from data/df_tile_coordinates.pkl to tile the WSIs with the coordinates used in our publication. In order to speed up this example, we sampled 2500 tiles at 40X without replacement for cases with more than 2500 tiles. Tiles will be saved to directories in path/to/tiles. n_jobs controls the number of parallel processes during tiling. Using too many jobs may result in a memory error, depending on your computing resources.
Fit models to predict gene expression in co-expression-based clusters by running
python train.py --indir path/to/tiles --modeldir path/to/models
Mode checkpoints will be saved under path/to/models.
train.py has several optional arguments to ensure that the code will run on your system:
--n_workersdetermines the number of workers in the data loader, all available workers will be used by default.--batch_sizedetermines the batch size per GPU, with a default of 100.--mixeddetermines whether to use mixed precision during model training and is deactivated by default.--multi_gpuactivates multi-gpu training if set toTrue, the default isFalse.
If you would like to change any further options such as hyperparameters, we suggest to directly edit train.py.
Navigate to 3_prediction and run
python predict.py --indir path/to/tiles --modeldir path/to/models
To predict on the outer CV fold and the test set. The optional parameter --n_workers controls the number of workers per pytorch dataloader, --batch_size controls the batch size per GPU. Tile-wise predictions per model will be saved under data/predictions_valid for the cross validation data and under data/predictions_test for the test set. Running
python pool_predictions_val.py
and
python pool_predictions_test.py
will average tile-wise predictions to WSI-level predictions and create the data frames data/df_preds_valid.pkl and data/df_preds_test.pkl which contain a prediction per expression level for each patient.
Under 4_analysis, there is an example of an analysis of the Spearman correlations in the validation and test data as conducted in our publication. All our model predictions and analysis results are publicly available under from Zenodo.