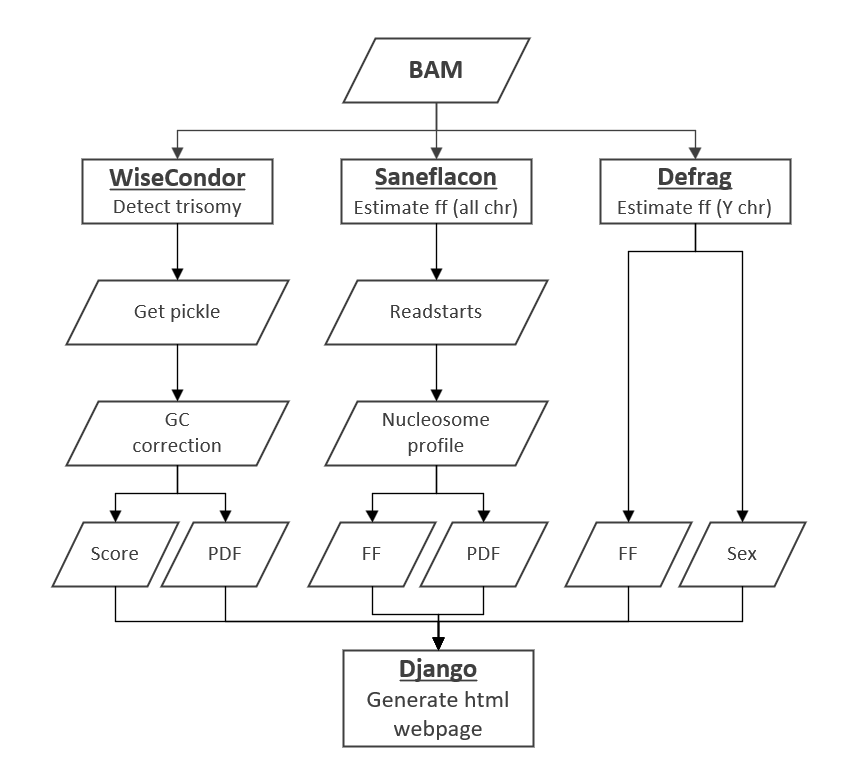
Tools used from other projects are shown below.
- Python
--pickle
--joblib
--scipy
--sklearn
Simply clone the repository:
git clone --recursive https://github.com/AllanSSX/IonNIPT.git
/!\ The first step is to train Defrag and Sanefalcon. Please, follow the documentation of each tool.
After that, you need to change some variables in IonNIPT.py:
- l.37: path to the trained model (Sanefalcon)
- l.40: the scaling factor value for Defrag
- l.109/113: number of jobs/threads. By default, total amount of cores for the Proton server.
- l.159 & 163: enable/disable the second module for sex determination. By default, disable.
In sanefalcon/predict.sh:
- l.2: path to predict.py
And if you using Torrent Server >= 5.2 and Ubuntu 14.04, please add the following correction in wisecondor/test.py:
- l.49:
if len(reference) == 0 or stddev == 0:
Moreover, you need to traine Sanefalcon and Defrag on your data and push the results into data/.
See Sanefalcon and Wisecondor manual for further details.
Structuration of the data folder:
data/
|__ female-defrag/
| |__ GRO-51_BRE00067.gcc
| |__ GRO-51_BRE00067.pickle
| |__ ...
| |__ GRO-71_BRE00127.gcc
| |__ GRO-71_BRE00127.pickle
|__ hg19.gccount
|__ male-defrag/
| |__ GRO-51_BRE00063.gcc
| |__ GRO-51_BRE00063.pickle
| |__ ...
| |__ GRO-72_BRE00128.gcc
| |__ GRO-72_BRE00128.pickle
|__ nuclTrack.1
|__ nuclTrack.10
|__ ...
|__ nuclTrack.8
|__ nuclTrack.9
|__ reftable
|__ trainModel.model
Just contact me, I will send you all the data
The aim of the function genesYspecificsSexDet() is to perform a second analysis to complete the Defrag test for sex determination is case of conflict between the sex cluster and the gender. It will calculate the percentage of reads for a subset of Y genes, specifics to the male (i.e. without orthologs in autosomes or in X). To activate the function, you need to uncomment l.159 & 163 from IonNIPT.py and fixe a threshold l.265 & 267 to determine the correct sex.
To identify these treshold values, simply run coverageYspecificGenes.py on your data for which the sex is already know and plot the result as shown in the next figure.