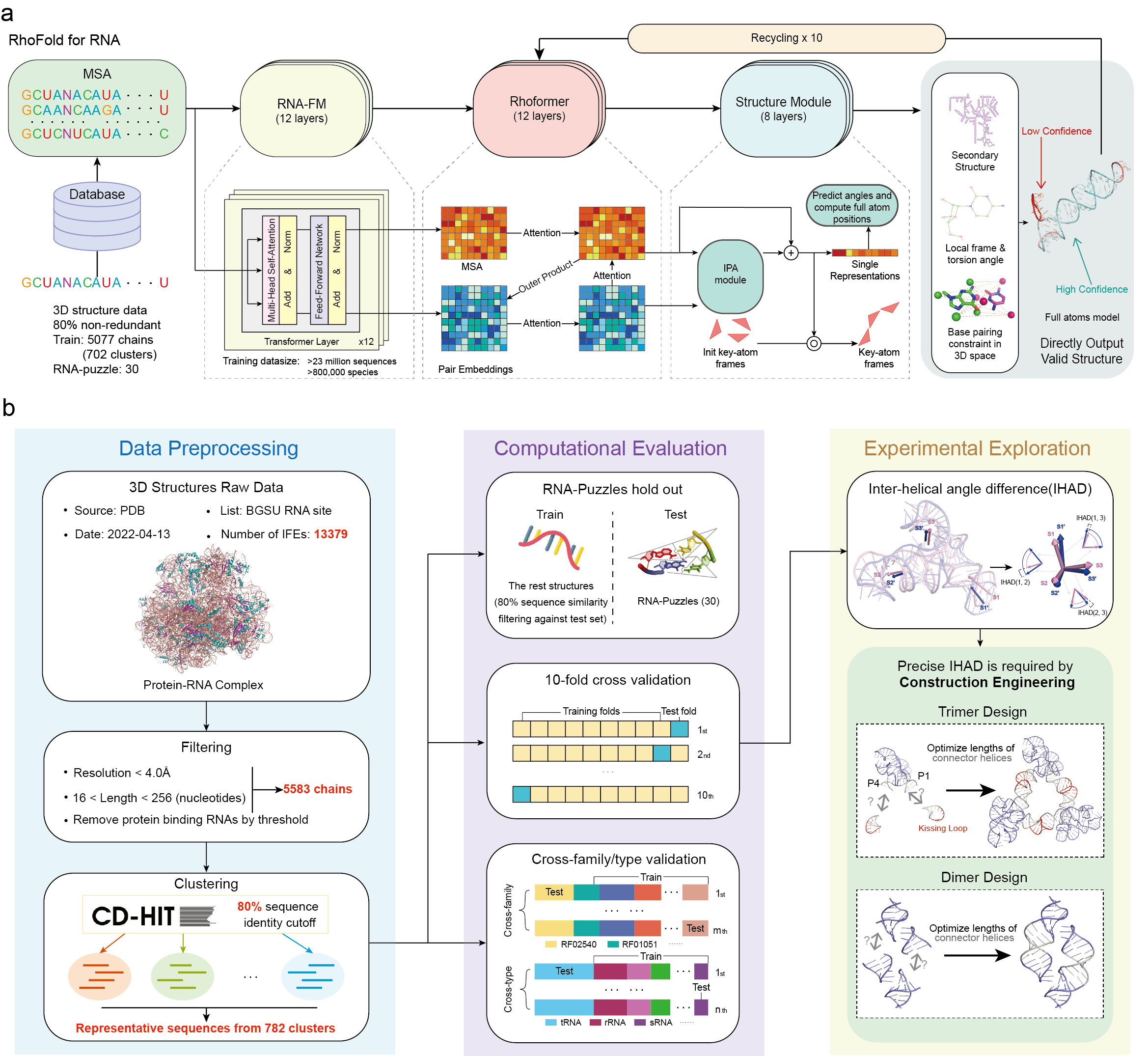
This is the opensource code for RhoFold.
Citation
@article{shen2022e2efold,
title={E2Efold-3D: End-to-End Deep Learning Method for accurate de novo RNA 3D Structure Prediction},
author={Shen, Tao and Hu, Zhihang and Peng, Zhangzhi and Chen, Jiayang and Xiong, Peng and Hong, Liang and Zheng, Liangzhen and Wang, Yixuan and King, Irwin and Wang, Sheng and others},
journal={arXiv preprint arXiv:2207.01586},
year={2022}
}Table of contents
10/10/2022
Initial commits:
- Pretrained model is provided.
No need to create the environment locally, you can also access RhoFold easily through its online server: https://proj.cse.cuhk.edu.hk/aihlab/rhofold/
Create Environment with Conda First, download the repository and create the environment.
(MacOS is currently not supported)
git clone https://github.com/RFOLD/RhoFold.git
cd ./RhoFold
conda env create -f ./envs/environment_linux.yaml
Then, activate the "rhofold" environment.
conda activate rhofold
python setup.py install
cd ./pretrained
wget https://proj.cse.cuhk.edu.hk/aihlab/rhofold/api/download?filename=rhofold_pretrained.pt -O rhofold_pretrained.pt
cd ../
python inference.py
--input_fas INPUT_FAS
Path to the input fasta file. Valid nucleic acids in RNA sequence: A, U, G, C. Input of sequence standalone is in testing. It's not as accurate as inputs of sequences combined with MSA. The former is only for the user to generate a quick reference structure.
--input_a3m INPUT_A3M
Path to the input msa file, default None.
If --input_a3m is not given (set to None), MSA will be generated automatically.
--output_dir OUTPUT_DIR
Path to the output dir.
Tertiary Structure prediction is saved in .pdb format (pLDDT score is recorded in the B-factor column).
Distogram prediction is saved in .npz format.
Secondary structure prediction is save in .ct format.
--device DEVICE
Default cpu. If GPUs are available, you can set --device cuda:<GPU_index> for faster prediction.
--ckpt CKPT
Path to the pretrained model. Default ./pretrained/model_20221010_params.pt
--relax_steps RELAX_STEPS
Num of steps for structure refinement, default 1000.
--single_seq_pred
Default False.
If --single_seq_pred is set to True, the modeling will run using single sequence only (input_fas)
--database_dpath
Path to the sequence database for MSA construction. Default ./database
--binary_dpath
Path to the executable. Default ./rhofold/data/bin
The outputs will be saved in the directory provided via the --output_dir flag of inference.py.
The outputs include the unrelaxed structures, relaxed structures, prediction metadata, and running log.
The --output_dir directory will have the following structure:
<--output_dir>/
results.npz
ss.ct
unrelaxed_model.pdb
relaxed_{relax_steps}_model.pdb
log.txt
The contents of each output file are as follows:
results.npz– A.npzfile containing the distogram prediction of RhoFold in NumPy arrays.ss.ct– A .ct format text file containing the predicted secondary structure.unrelaxed_model.pdb– A PDB format file containing the predicted structure from deep learning.relaxed_{relax_steps}_model.pdb– A PDB format file containing the amber relaxed structure from unrelaxed_model.pdb.log.txt– A txt file containing the running log.
Below are examples on how to use RhoFold in different scenarios.
python inference.py --input_fas ./example/input/3owzA/3owzA.fasta --input_a3m ./example/input/3owzA/3owzA.a3m --output_dir ./example/output/3owzA/ --ckpt ./pretrained/rhofold_pretrained.pt
1.Sequence standalone
This function is in testing. It's not as accurate as the MSA version. It's only for the user to generate a quick reference structure.
python inference.py --input_fas ./example/input/3owzA/3owzA.fasta --single_seq_pred True --output_dir ./example/output/3owzA/ --ckpt ./pretrained/rhofold_pretrained.pt
2.With our constructed MSA (Full version of RhoFold)
To support MSA construction, 3 sequence databases (RNAcentral, Rfam, and nt) totaling about 900GB need to be downloaded.
Warning: you should ensure that there are adequate spaces for saving the data! Otherwise you can directly utilize our online server, or download our off-the-shelf MSAs instead of regenerating them.
./database/bin/builddb.sh
Then you can run the following command lines:
python inference.py --input_fas ./example/input/3owzA/3owzA.fasta --output_dir ./example/output/3owzA/ --ckpt ./pretrained/rhofold_pretrained.pt
You can access training data (13.86G) from the google drive link. The file includes the off-the-shelf MSAs of training data, which can be fed into RhoFold directly.
@article{shen2022e2efold,
title={E2Efold-3D: End-to-End Deep Learning Method for accurate de novo RNA 3D Structure Prediction},
author={Shen, Tao and Hu, Zhihang and Peng, Zhangzhi and Chen, Jiayang and Xiong, Peng and Hong, Liang and Zheng, Liangzhen and Wang, Yixuan and King, Irwin and Wang, Sheng and others},
journal={arXiv preprint arXiv:2207.01586},
year={2022}
}This source code is licensed under the Apache license found in the LICENSE file
in the root directory of this source tree.