QuickAnnotator
Quick Annotator is an open-source digital pathology annotation tool.
Purpose
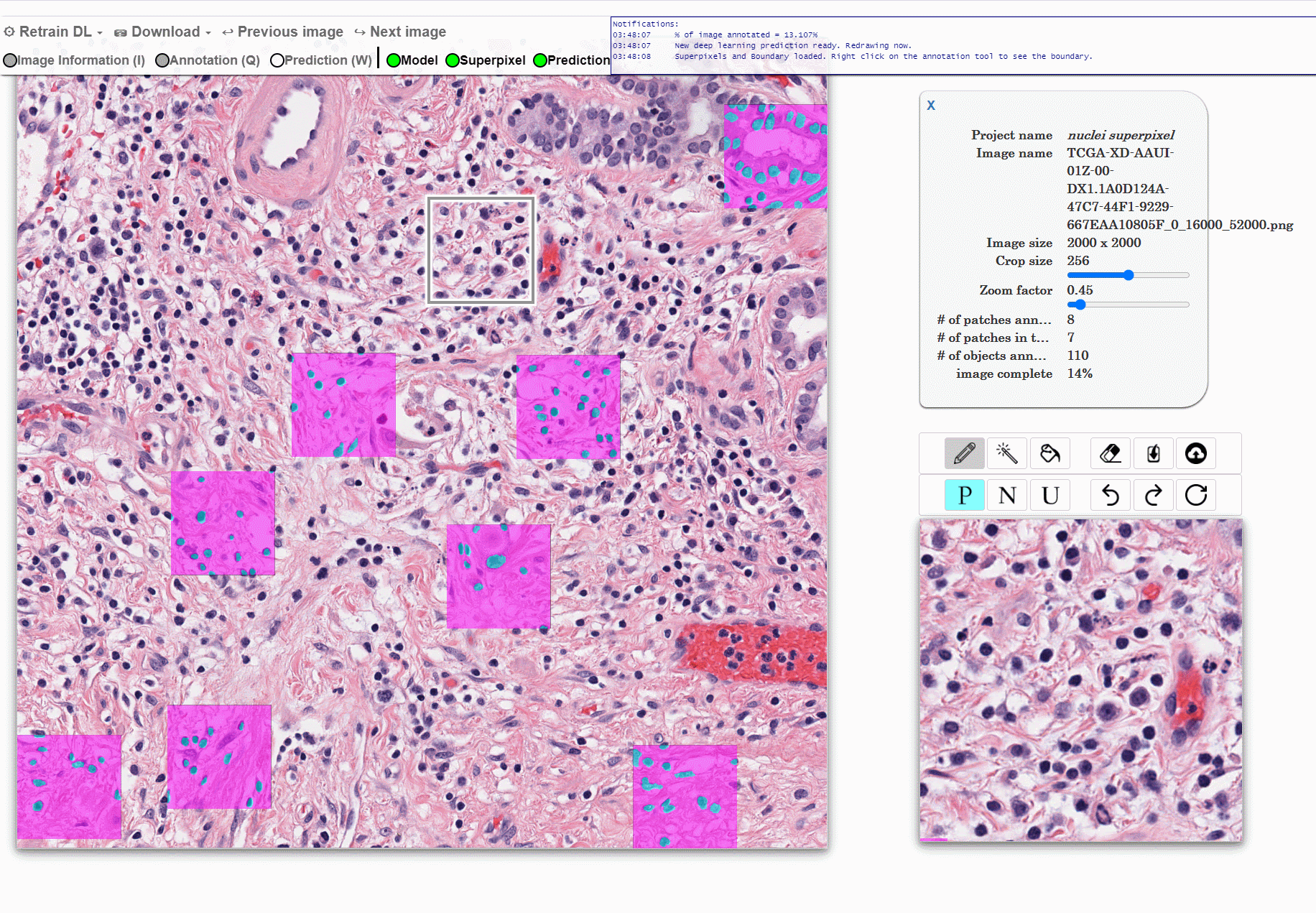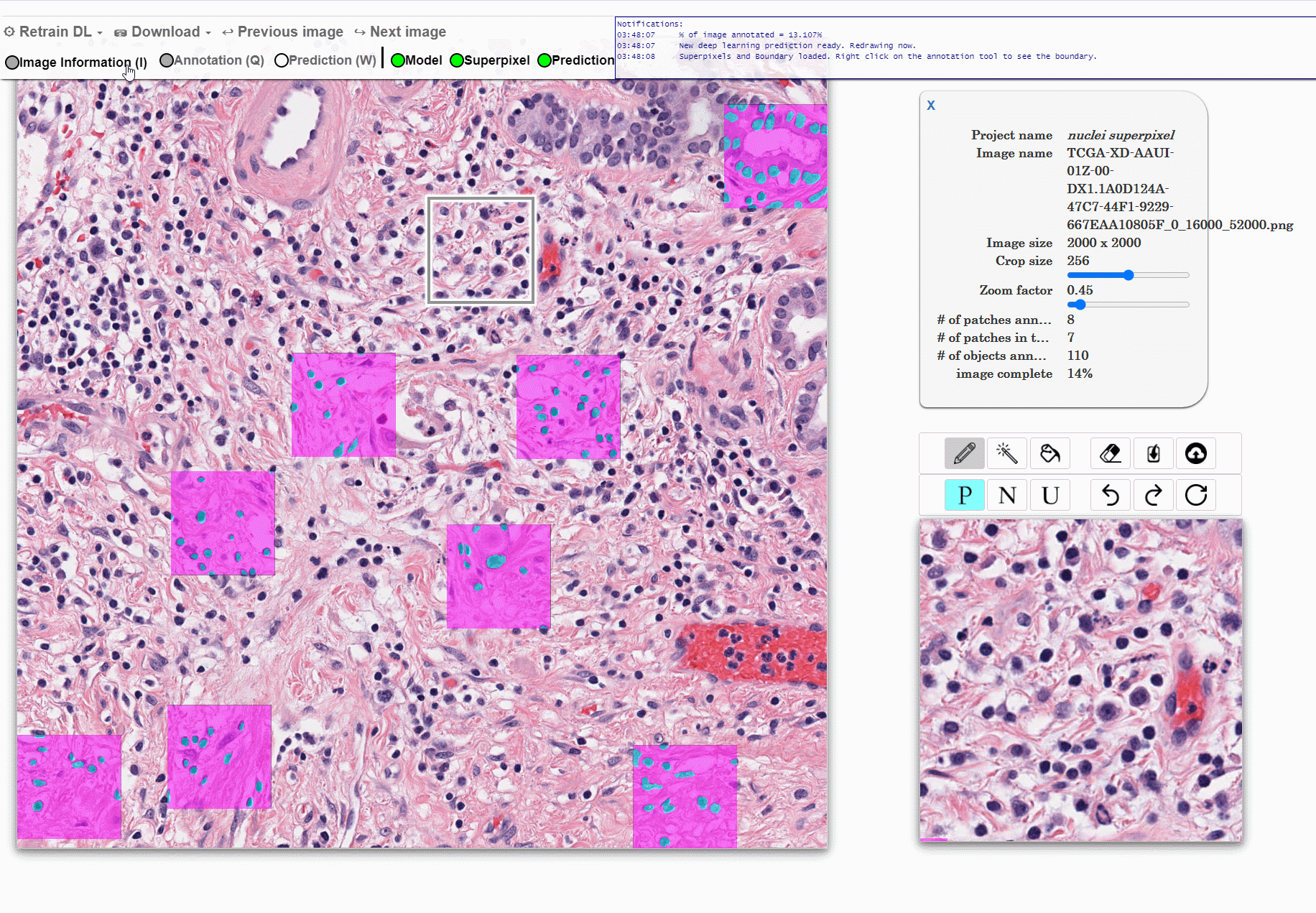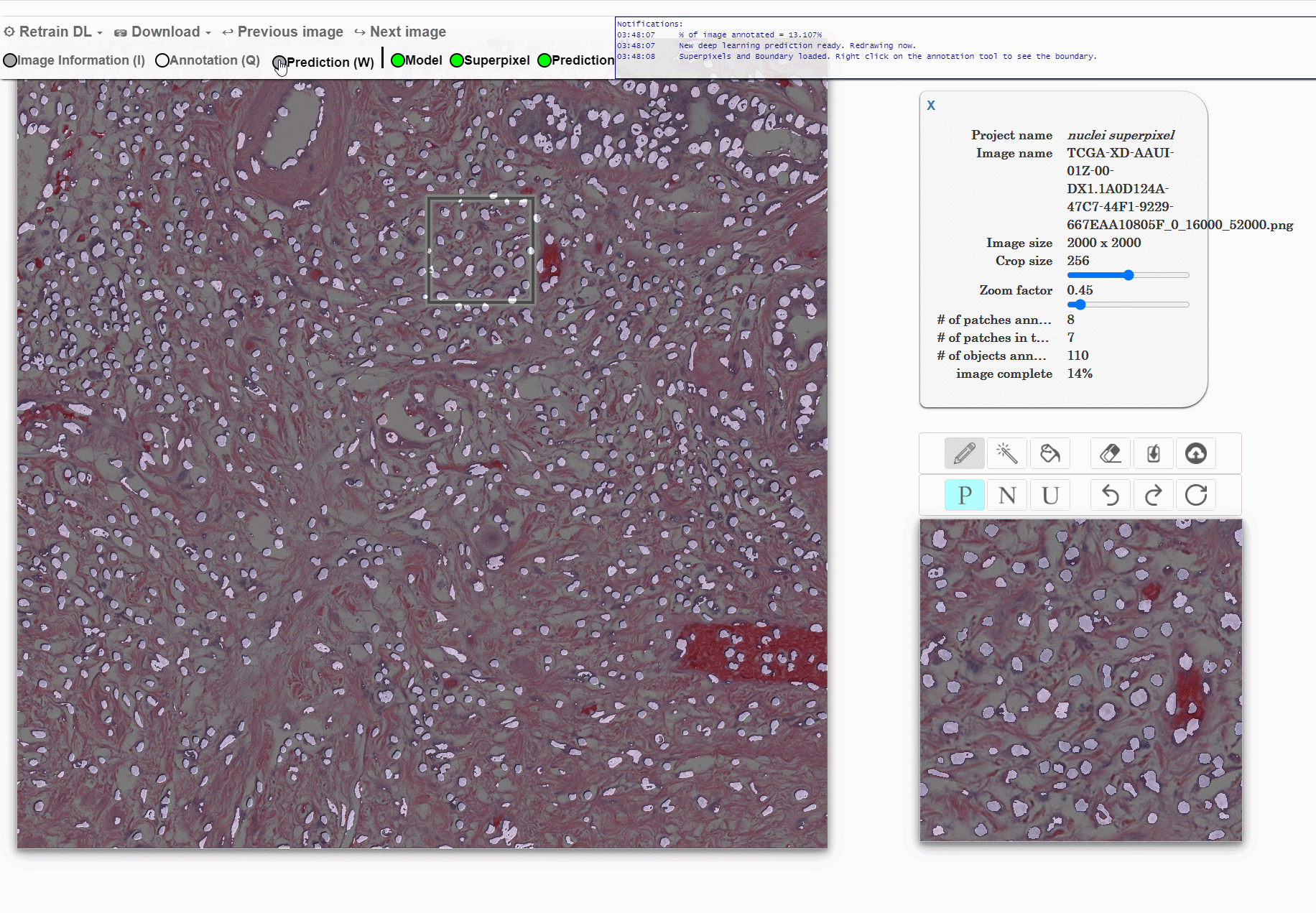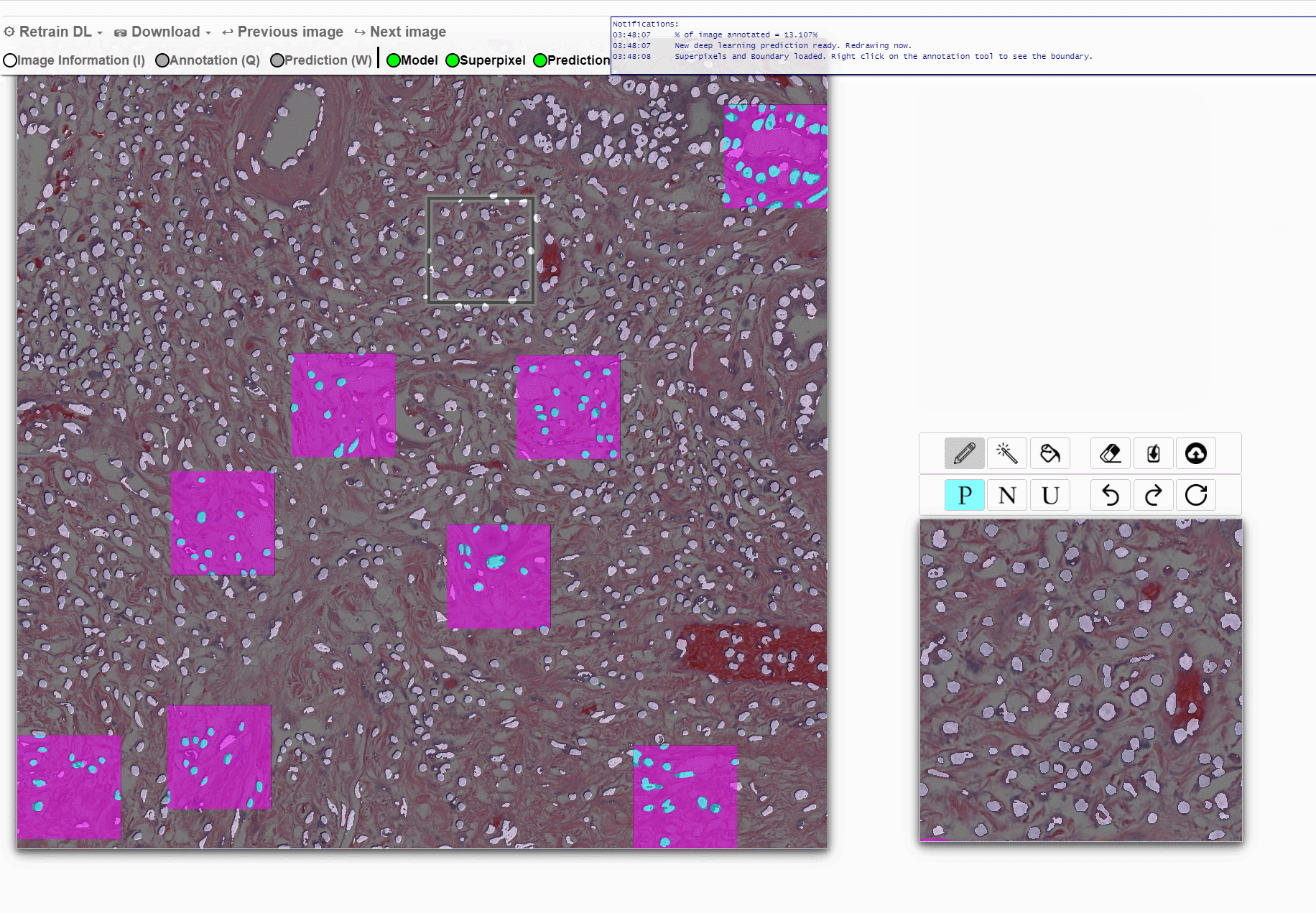
Machine learning approaches for segmentation of histologic primitives (e.g., cell nuclei) in digital pathology (DP) Whole Slide Images (WSI) require large numbers of exemplars. Unfortunately, annotating each object is laborious and often intractable even in moderately sized cohorts. The purpose of the quick annotator is to rapidly bootstrap annotation creation for digital pathology projects by helping identify images and small regions.
Because the classifier is likely to struggle by intentionally focusing in these areas, less pixel level annotations are needed, which significantly improves the efficiency of the users. Our approach involves updating a u-net model while the user provides annotations, so the model is then in real time used to produce. This allows the user to either accept or modify regions of the prediction.
Requirements
Tested with Python 3.8
Requires:
- Python
- pip
And the following additional python package:
- Flask_SQLAlchemy
- scikit_image
- scikit_learn
- opencv_python_headless
- scipy
- requests
- SQLAlchemy
- torch
- torchvision 10.Flask_Restless
- numpy
- Flask
- umap_learn
- Pillow
- tensorboardX
- ttach
- albumentations
- config
You can likely install the python requirements using something like (note python 3+ requirement):
pip3 install -r requirements.txt
The library versions have been pegged to the current validated ones. Later versions are likely to work but may not allow for cross-site/version reproducibility
We received some feedback that users could installed torch. Here, we provide a detailed guide to install Torch
Torch's Installation
The general guides for installing Pytorch can be summarized as following:
- Check your NVIDIA GPU Compute Capability @ https://developer.nvidia.com/cuda-gpus
- Download CUDA Toolkit @ https://developer.nvidia.com/cuda-downloads
- Install PyTorch command can be found @ https://pytorch.org/get-started/locally/
Basic Usage
see UserManual for a demo
Run
E:\Study\Research\QA\qqqqq\test1\quick_annotator>python QA.py
By default, it will start up on localhost:5555
Warning: virtualenv will not work with paths that have spaces in them, so make sure the entire path to env/ is free of spaces.
Config Sections
There are many modular functions in QA whose behaviors could be adjusted by hyper-parameters. These hyper-parameters can be set in the config.ini file
- [common]
- [flask]
- [cuda]
- [sqlalchemy]
- [pooling]
- [train_ae]
- [train_tl]
- [make_patches]
- [make_embed]
- [get_prediction]
- [frontend]
- [superpixel]
Advanced Usage
See wiki
Citation
Read the related paper on arXiv: Quick Annotator: an open-source digital pathology based rapid image annotation tool
PDF file available for download
Please use below to cite this paper if you find this repository useful or if you use the software shared here in your research.
@misc{miao2021quick,
title={Quick Annotator: an open-source digital pathology based rapid image annotation tool},
author={Runtian Miao and Robert Toth and Yu Zhou and Anant Madabhushi and Andrew Janowczyk},
year={2021},
eprint={2101.02183},
archivePrefix={arXiv},
primaryClass={eess.IV}
}
Frequently Asked Questions
See FAQ